Comparative Genomic Analyses of Flavobacterium psychrophilum Isolates Reveals New Putative Genetic Determinants of Virulence Traits
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain Isolation, Medium Composition, and Growth Conditions
2.2. DNA Extraction
2.3. Genome Sequencing, Assembly, and Annotation
2.4. Predictions of Genomic Islands, Virulence-Related Factors and Prophages
2.5. Pan Genome Analysis
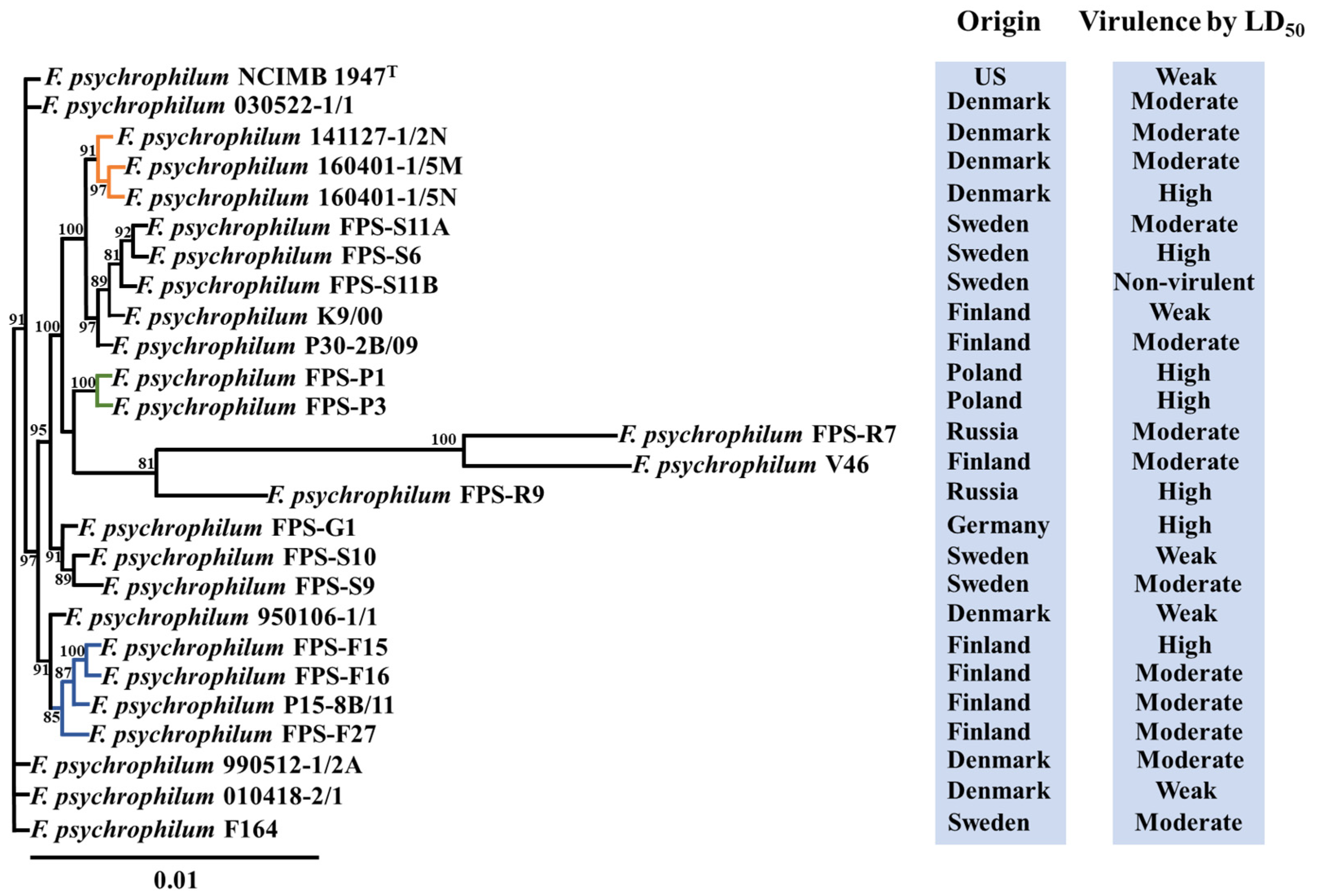
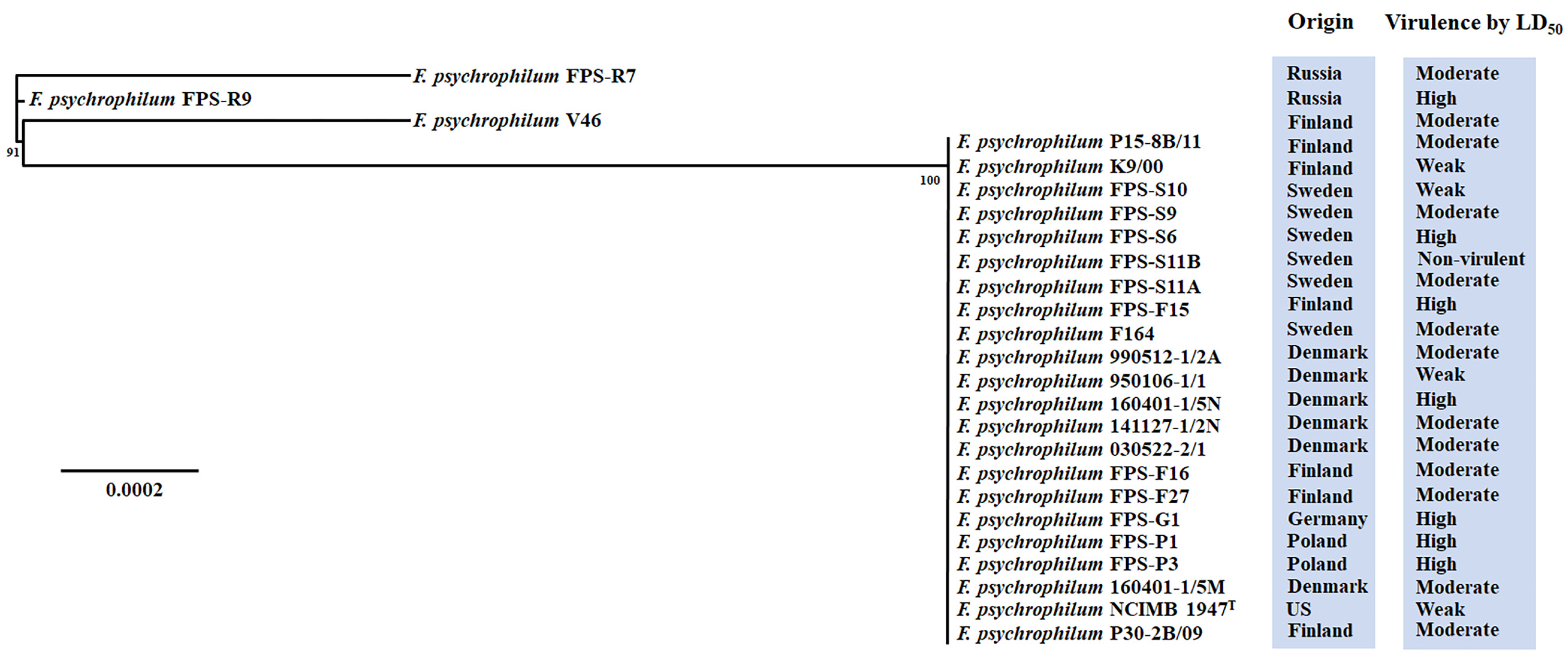
2.6. Phylogenetic Analysis
2.7. Accession Numbers
3. Results
3.1. Virulence Properties of F. psychrophilum Isolates
3.2. Genomic Characteristics of F. psychrophilum Isolates
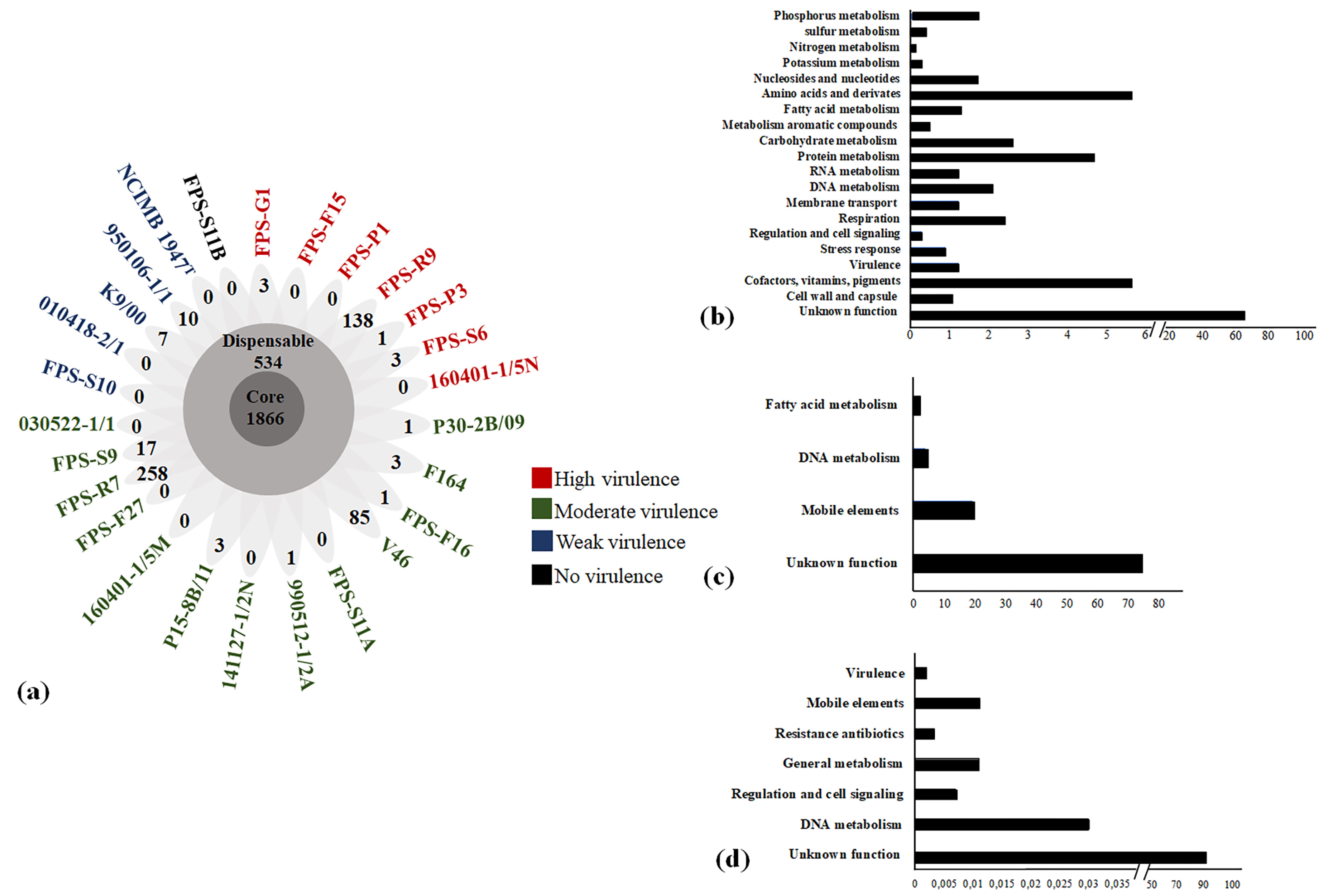
3.3. Relation of F. psychrophilum Pan Genome and Virulence Traits
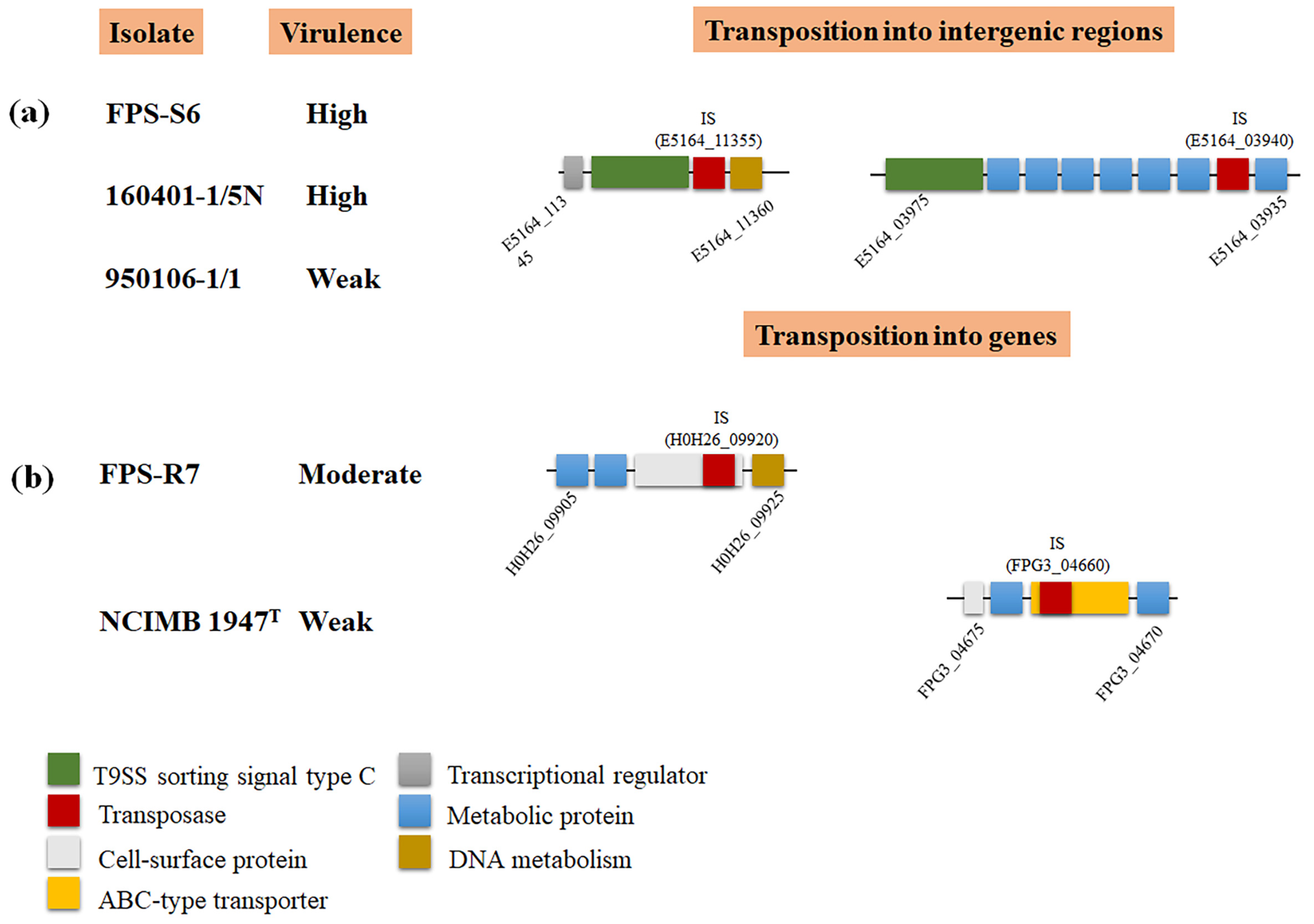
3.4. Precision Long-Read Sequencing for F. psychrophilum
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nematollahi, A.; Decostere, A.; Pasmans, F.; Haesebrouck, F. Flavobacterium psychrophilum infections in salmonid fish. J. Fish Dis. 2003, 26, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Barnes, M.E.; Brown, M.L. A review of Flavobacterium psychrophilum biology, clinical signs, and bacterial cold water disease prevention and treatment. Open Fish Sci. J. 2011, 4, 40–48. [Google Scholar] [CrossRef]
- Duchaud, E.; Boussaha, M.; Loux, V.; Bernardet, J.F.; Michel, C.; Kerouault, B.; Mondot, S.; Nicolas, P.; Bossy, R.; Caron, C.; et al. Complete genome sequence of the fish pathogen Flavobacterium psychrophilum. Nat. Biotechnol. 2007, 25, 763–769. [Google Scholar] [CrossRef] [PubMed]
- Sundell, K.; Landor, L.; Nicolas, P.; Jørgensen, J.; Castillo, D.; Middelboe, M.; Dalsgaard, I.; Donati, V.L.; Madsen, L.; Wiklund, T. Phenotypic and genetic predictors of pathogenicity and virulence in Flavobacterium psychrophilum. Front. Microbiol. 2019, 10, 1711. [Google Scholar] [CrossRef]
- Högfors-Rönnholm, E.; Wiklund, T. Hemolytic activity in Flavobacterium psychrophilum is a contact-dependent, two-step mechanism and differently expressed in smooth and rough phenotypes. Microb. Pathog. 2010, 49, 369–375. [Google Scholar] [CrossRef]
- Castillo, D.; Christiansen, R.H.; Dalsgaard, I.; Madsen, L.; Middelboe, M. Bacteriophage resistance mechanisms in the fish pathogen Flavobacterium psychrophilum: Linking genomic mutations to changes in bacterial virulence factors. Appl. Environ. Microbiol. 2015, 81, 1157–1167. [Google Scholar] [CrossRef]
- Barbier, P.; Rochat, T.; Mohammed, H.H.; Wiens, G.D.; Bernardet, J.F.; Halpern, D.; Duchaud, E.; McBride, M.J. The type IX secretion system is required for virulence of the fish pathogen Flavobacterium psychrophilum. Appl. Environ. Microbiol. 2020, 86, e00799–e00820. [Google Scholar] [CrossRef]
- Duchaud, E.; Rochat, T.; Habib, C.; Barbier, P.; Loux, V.; Guérin, C.; Dalsgaard, I.; Madsen, L.; Nilsen, H.; Sundell, K.; et al. Genomic diversity and evolution of the fish pathogen Flavobacterium psychrophilum. Front. Microbiol. 2018, 9, 138. [Google Scholar] [CrossRef]
- Nilsen, H.; Sundell, K.; Duchaud, E.; Nicolas, P.; Dalsgaard, I.; Madsen, L.; Aspán, A.; Jansson, E.; Colquhoun, D.J.; Wiklund, T. Multilocus sequence typing identifies epidemic clones of Flavobacterium psychrophilum in Nordic countries. Appl. Environ. Microbiol. 2014, 80, 2728–2736. [Google Scholar] [CrossRef]
- Castillo, D.; Christiansen, R.H.; Espejo, R.; Middelboe, M. Diversity and geographical distribution of Flavobacterium psychrophilum isolates and their phages: Patterns of susceptibility to phage infection and phage host range. Microb. Ecol. 2014, 67, 748–757. [Google Scholar] [CrossRef]
- Castillo, D.; Christiansen, R.H.; Dalsgaard, I.; Madsen, L.; Espejo, R.; Middelboe, M. Comparative genome analysis provides insights into the pathogenicity of Flavobacterium psychrophilum. PLoS ONE 2016, 11, e0152515. [Google Scholar] [CrossRef]
- Castillo, D.; Jørgensen, J.; Sundell, K.; Madsen, L.; Dalsgaard, I.; Wiklund, T.; Middelboe, M. Genome-informed approach to identify genetic determinants of Flavobacterium psychrophilum phage susceptibility. Environ. Microbiol 2021, in press. [Google Scholar]
- Pérez-Pascual, D.; Rochat, T.; Kerouault, B.; Gómez, E.; Neulat-Ripoll, F.; Henry, C.; Quillet, E.; Guijarro, J.A.; Bernardet, J.F.; Duchaud, E. More than gliding: Involvement of GldD and GldG in the virulence of Flavobacterium psychrophilum. Front. Microbiol. 2017, 8, 2168. [Google Scholar] [CrossRef]
- Stenholm, A.R.; Dalsgaard, I.; Middelboe, M. Isolation and characterization of bacteriophages infecting the fish pathogen Flavobacterium psychrophilum. Appl. Environ. Microbiol. 2008, 74, 4070–4078. [Google Scholar] [CrossRef]
- Castillo, D.; Higuera, G.; Villa, M.; Middelboe, M.; Dalsgaard, I.; Madsen, L.; Espejo, R.T. Diversity of Flavobacterium psychrophilum and the potential use of its phages for protection against bacterial cold water disease in salmonids. J. Fish Dis. 2012, 35, 193–201. [Google Scholar] [CrossRef]
- Donati, V.L.; Dalsgaard, I.; Sundell, K.; Castillo, D.; Er-Rafik, M.; Clark, J.; Wiklund, T.; Middelboe, M.; Madsen, L. Phage-mediated control of Flavobacterium psychrophilum in aquaculture: In vivo experiments to compare delivery methods. Front. Microbiol. 2021, 12, 628309. [Google Scholar] [CrossRef] [PubMed]
- Castillo, D.; Middelboe, M. Genomic diversity of bacteriophages infecting the fish pathogen Flavobacterium psychrophilum. FEMS Microbiol. Lett. 2016, 363, fnw272. [Google Scholar] [CrossRef][Green Version]
- Gyles, C.; Boerlin, P. Horizontally transferred genetic elements and their role in pathogenesis of bacterial disease. Vet. Pathol. 2014, 51, 328–340. [Google Scholar] [CrossRef] [PubMed]
- Cerveny, L.; Straskova, A.; Dankova, V.; Hartlova, A.; Ceckova, M.; Staud, F.; Stulik, J. Tetratricopeptide repeat motifs in the world of bacterial pathogens: Role in virulence mechanisms. Infect. Immun. 2013, 81, 629–635. [Google Scholar] [CrossRef]
- Bliven, K.A.; Maurelli, A.T. Antivirulence genes: Insights into pathogen evolution through gene loss. Infect. Immun. 2012, 80, 4061–4070. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.K.; Kropinski, A.M.; Lumsden, J.S.; Dixon, B.; MacInnes, J.I. Complete genome sequence of the fish pathogen Flavobacterium psychrophilum ATCC 49418(T.). Stand. Genomic Sci. 2015, 10, 3. [Google Scholar] [CrossRef]
- Holt, R.A.; Rohovec, J.S.; Fryer, J.L. Bacterial cold-water disease. In Bacterial Disease of Fish, 1st ed.; Inglis, V., Roberts, R.J., Bromage, N.R., Eds.; Blackwell Scientific Publications: Oxford, UK, 1993; pp. 3–22. [Google Scholar]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Seppey, M.; Manni, M.; Zdobnov, E.M. BUSCO: Assessing Genome Assembly and Annotation Completeness. In Gene Prediction. Methods in Molecular Biology, 1st ed.; Kollmar, M., Ed.; Spring Protocols: New York, NY, USA, 2019; Volume 1962, pp. 227–245. [Google Scholar]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Ruiqiang, L.; Yingrui, L.; Karsten, K.; Jun, W. SOAP: Short oligonucleotide alignment program. Bioinformatics 2008, 24, 713–714. [Google Scholar]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef] [PubMed]
- Bertelli, C.; Laird, M.R.; Williams, K.P.; Simon Fraser University Research Computing Group; Lau, B.Y.; Hoad, G.; Winsor, G.L.; Brinkman, F.S.L. IslandViewer 4: Expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017, 45, W30–W35. [Google Scholar] [CrossRef]
- Darling, A.C.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef]
- Zhou, C.E.; Smith, J.; Lam, M.; Zemla, A.; Dyer, M.D.; Slezak, T. MvirDB a microbial database of protein toxins, virulence factors and antibiotic resistance genes for bio-defence applications. Nucleic Acids Res. 2007, 35, D391–D394. [Google Scholar] [CrossRef] [PubMed]
- Kleinheinz, K.A.; Joensen, K.G.; Larsen, M.V. Applying the ResFinder and VirulenceFinder web-services for easy identification of acquired antibiotic resistance and E. coli virulence genes in bacteriophage and prophage nucleotide sequences. Bacteriophage 2014, 4, e27943. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Arndt, D.; Marcu, A.; Liang, Y.; Wishart, D.S. Phast, phaster and phastest: Tools for finding prophage in bacterial genomes. Brief. Bioinform. 2019, 20, 1560–1567. [Google Scholar] [CrossRef]
- Yu, N.Y.; Wagner, J.R.; Laird, M.R.; Melli, G.; Rey, S.; Lo, R.; Dao, P.; Sahinalp, S.C.; Ester, M.; Foster, L.J.; et al. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 2010, 26, 1608–1615. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Emanuelsson, O.; Brunak, S.; von Heijne, G.; Nielsen, H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007, 2, 953–971. [Google Scholar] [CrossRef] [PubMed]
- Pagni, M.; Ioannidis, V.; Cerutti, L.; Zahn-Zabal, M.; Jongeneel, C.V.; Hau, J.; Martin, O.; Kuznetsov, D.; Falquet, L. MyHits: Improvements to an interactive resource for analyzing protein sequences. Nucleic Acids Res. 2007, 35, W433–W437. [Google Scholar] [CrossRef] [PubMed]
- Blom, J.; Kreis, J.; Spänig, S.; Juhre, T.; Bertelli, C.; Ernst, C.; Goesmann, A. EDGAR 2.0: An enhanced software platform for comparative gene content analyses. Nucleic Acids Res. 2016, 44, W22–W28. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Mackey, A.J.; Stoeckert, C.J., Jr.; Roos, D.S. OrthoMCL-DB: Querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006, 34, D363–D368. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Kück, P.; Meusemann, K. FASconCAT: Convenient handling of data matrices. Mol. Phylogenetics Evol. 2010, 56, 1115–1118. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Álvarez, B.; Álvarez, J.; Menéndez, A.; Guijarro, J.A. A mutant in one of two exbD loci of a TonB system in Flavobacterium psychrophilum shows attenuated virulence and confers protection against cold water disease. Microbiology 2008, 154, 1144–1151. [Google Scholar] [CrossRef]
- Nakayama, H.; Tanaka, K.; Teramura, N.; Hattori, S. Expression of collagenase in Flavobacterium psychrophilum isolated from cold-water disease-affected ayu (Plecoglossus altivelis). Biosci. Biotechnol. Biochem. 2016, 80, 135–144. [Google Scholar] [CrossRef]
- Pérez-Pascual, D.; Gómez, E.; Guijarro, J.A. Lack of a type-2 glycosyltransferase in the fish pathogen Flavobacterium psychrophilum determines pleiotropic changes and loss of virulence. Vet. Res. 2015, 46, 1–9. [Google Scholar] [CrossRef]
- Alvarez, B.; Secades, P.; Prieto, M.; McBride, M.J.; Guijarro, J.A. A mutation in Flavobacterium psychrophilum tlpB inhibits gliding motility and induces biofilm formation. Appl. Environ. Microbiol. 2006, 72, 4044–4053. [Google Scholar] [CrossRef]
- Castillo, D.; Espejo, R.; Middelboe, M. Genomic structure of bacteriophage 6H and its distribution as prophage in Flavobacterium psychrophilum strains. FEMS Microbiol. Lett. 2014, 351, 51–58. [Google Scholar] [CrossRef]
- Loman, N.J.; Quick, J.; Simpson, J.T. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat. Methods 2015, 12, 733–735. [Google Scholar] [CrossRef] [PubMed]
- Sundell, K.; Wiklund, T. Characteristics of epidemic and sporadic Flavobacterium psychrophilum sequence types. Aquaculture 2015, 441, 51–56. [Google Scholar] [CrossRef]
- Leitão, J.H. Microbial Virulence Factors. Int. J. Mol. Sci. 2020, 21, 5320. [Google Scholar] [CrossRef]
- Diard, M.; Hardt, W.D. Evolution of bacterial virulence. FEMS Microbiol. Rev. 2017, 41, 679–697. [Google Scholar] [CrossRef] [PubMed]
- Rouli, L.; Merhej, V.; Fournier, P.E.; Raoult, D. The bacterial pangenome as a new tool for analysing pathogenic bacteria. New Microbes New Infect. 2015, 7, 72–85. [Google Scholar] [CrossRef]
- Kim, Y.; Gu, C.; Kim, H.U.; Lee, S.Y. Current status of pan-genome analysis for pathogenic bacteria. Curr. Opin. Biotechnol. 2020, 63, 54–62. [Google Scholar] [CrossRef]
- Castillo, D.; Alvise, P.D.; Xu, R.; Zhang, F.; Middelboe, M.; Gram, L. Comparative genome analyses of Vibrio anguillarum strains reveal a link with pathogenicity traits. Msystems 2017, 28, e00001-17. [Google Scholar] [CrossRef] [PubMed]
- Frans, I.; Michiels, C.W.; Bossier, P.; Willems, K.A.; Lievens, B.; Rediers, H. Vibrio anguillarum as a fish pathogen: Virulence factors, diagnosis and prevention. J. Fish Dis. 2011, 34, 643–661. [Google Scholar] [CrossRef] [PubMed]
- Kaur, K.; Sharma, A.; Capalash, N.; Sharma, P. Multicopper oxidases: Biocatalysts in microbial pathogenesis and stress management. Microbiol. Res. 2019, 222, 1–13. [Google Scholar] [CrossRef]
- Stockbauer, K.E.; Magoun, L.; Liu, M.; Burns, E.H., Jr.; Gubba, S.; Renish, S.; Pan, X.; Bodary, S.C.; Baker, E.; Coburn, J.; et al. A natural variant of the cysteine protease virulence factor of group A Streptococcus with an arginine-glycine-aspartic acid (RGD) motif preferentially binds human integrins alphavbeta3 and alphaIIbbeta3. Proc. Natl. Acad. Sci. USA 1999, 96, 242–247. [Google Scholar] [CrossRef]
- Gharrah, M.M.; Mostafa El-Mahdy, A.; Barwa, R.F. Association between virulence factors and extended spectrum beta-lactamase producing Klebsiella pneumoniae compared to nonproducing isolates. Interdiscip. Perspect. Infect. Dis. 2017, 2017, 7279830. [Google Scholar] [CrossRef]
- Alcalde-Rico, M.; Hernando-Amado, S.; Blanco, P.; Martínez, J.L. Multidrug efflux pumps at the crossroad between antibiotic resistance and bacterial virulence. Front. Microbiol. 2016, 7, 1483. [Google Scholar] [CrossRef]
- Vandecraen, J.; Chandler, M.; Aertsen, A.; Van Houdt, R. The impact of insertion sequences on bacterial genome plasticity and adaptability. Crit. Rev. Microbiol. 2017, 43, 709–730. [Google Scholar] [CrossRef] [PubMed]
- Rojo-Bezares, B.; Estepa, V.; Cebollada, R.; de Toro, M.; Somalo, S.; Seral, C.; Castillo, F.J.; Torres, C.; Saenz, Y. Carbapenem resistant Pseudomonas aeruginosa strains from a Spanish hospital: Characterization of metallo-beta-lactamases, porin OprD and integrons. Int. J. Med. Microbiol. 2014, 304, 405–414. [Google Scholar] [CrossRef]
- Benson, M.A.; Ohneck, E.A.; Ryan, C.; Alonzo III, F.; Smith, H.; Narechania, A.; Kolokotronis, S.O.; Satola, S.W.; Uhlemann, A.C.; Sebra, R.; et al. Evolution of hypervirulence by a MRSA clone through acquisition of a transposable element. Mol. Microbiol. 2014, 93, 664–681. [Google Scholar] [CrossRef]
- Christie-Oleza, J.A.; Nogales, B.; Martin-Cardona, C.; Lanfranconi, M.P.; Alberti, S.; Lalucat, J.; Bosch, R. ISPst9, an ISL3-like insertion sequence from Pseudomonas stutzeri AN10 involved in catabolic gene inactivation. Int. Microbiol. 2008, 11, 101–110. [Google Scholar]
- Arciola, C.R.; Campoccia, D.; Ravaioli, S.; Montanaro, L. Polysaccharide intercellular adhesin in biofilm: Structural and regulatory aspects. Front. Cell. Infect. Microbiol. 2015, 5, 7. [Google Scholar] [CrossRef]
- Perez, M.; Calles-Enriquez, M.; del Rio, B.; Ladero, V.; Martin, M.C.; Fernandez, M.; Alvarez, M.A. IS256 abolishes gelatinase activity and biofilm formation in a mutant of the nosocomial pathogen Enterococcus faecalis V583. Can. J. Microbiol. 2015, 61, 517–519. [Google Scholar] [CrossRef]
- Graves, J.L., Jr.; Tajkarimi, M.; Cunningham, Q.; Campbell, A.; Nonga, H.; Harrison, S.H.; Barrick, J.E. Rapid evolution of silver nanoparticle resistance in Escherichia coli. Front. Genet. 2015, 6, 42. [Google Scholar] [CrossRef]
- Sun, X.; Dennis, J.J. A novel insertion sequence derepresses efflux pump expression and preadapts Pseudomonas putida S12 for extreme solvent stress. J. Bacteriol. 2009, 191, 6773–6777. [Google Scholar] [CrossRef]
- Cannatelli, A.; Giani, T.; D’Andrea, M.M.; Di Pilato, V.; Arena, F.; Conte, V.; Tryfinopoulou, K.; Vatopoulos, A.; Rossolini, G.M.; Group CS. MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob. Agents Chemother 2014, 58, 5696–5703. [Google Scholar] [CrossRef]
- Boutoille, D.; Corvec, S.; Caroff, N.; Giraudeau, C.; Espaze, E.; Caillon, J.; Plésiat, P.; Reynaud, A. Detection of an IS21 insertion sequence in the mexR gene of Pseudomonas aeruginosa increasing β-lactam resistance. FEMS Microbiol. Lett. 2004, 230, 143–146. [Google Scholar] [CrossRef]
- Kajava, A.V. Tandem repeats in proteins: From sequence to structure. J. Struct. Biol. 2012, 179, 279–288. [Google Scholar] [CrossRef]
- Kedzierski, Ł.; Montgomery, J.; Curtis, J.; Handman, E. Leucine-rich repeats in host-pathogen interactions. Arch. Immunol. Exp. 2004, 52, 104–112. [Google Scholar]
- Parida, S.K.; Domann, E.; Rohde, M.; Muller, S.; Darji, A.; Hain, T.; Wehland, J.; Chakraborty, T. Internalin B is essential for adhesion and mediates the invasion of Listeria monocytogenes into human endothelial cells. Mol. Microbiol. 1998, 28, 81–93. [Google Scholar] [CrossRef]
- Haraga, A.; Miller, S.I. A Salmonella enterica serovar typhimurium translocated leucine-rich repeat effector protein inhibits NF-κB-dependent gene expression. Infect. Immun. 2003, 71, 4052–4058. [Google Scholar] [CrossRef]
- Loimaranta, V.; Hytönen, J.; Pulliainen, A.T.; Sharma, A.; Tenovuo, J.; Strömberg, N.; Finne, J. Leucine-rich repeats of bacterial surface proteins serve as common pattern recognition motifs of human scavenger receptor gp340. J. Biol. Chem. 2009, 284, 18614–18623. [Google Scholar] [CrossRef]
- Brinster, S.; Posteraro, B.; Bierne, H.; Alberti, A.; Makhzami, S.; Sanguinetti, M.; Serror, P. Enterococcal leucine-rich repeat-containing protein involved in virulence and host inflammatory response. Infect. Immun. 2007, 75, 4463–4471. [Google Scholar] [CrossRef]
- Marino, M.; Braun, L.; Cossart, P.; Ghosh, P. Structure of the lnlB leucine-rich repeats, a domain that triggers host cell invasion by the bacterial pathogen L. monocytogenes. Mol. Cell 1999, 4, 1063–1072. [Google Scholar] [CrossRef]
- McBride, M.J.; Zhu, Y. Gliding motility and Por secretion system genes are widespread among members of the phylum Bacteroidetes. J. Bacteriol. 2013, 195, 270–278. [Google Scholar] [CrossRef]
- Lasica, A.M.; Ksiazek, M.; Madej, M.; Potempa, J. The type IX secretion system (T9SS): Highlights and recent insights into its structure and function. Front. Cell. Infect. Microbiol. 2017, 7, 215. [Google Scholar] [CrossRef]
- Rhodes, R.G.; Nelson, S.S.; Pochiraju, S.; McBride, M.J. Flavobacterium johnsoniae sprB is part of an operon spanning the additional gliding motility genes sprC, sprD, and sprF. J. Bacteriol. 2011, 193, 599–610. [Google Scholar] [CrossRef]
- Sato, K.; Naito, M.; Yukitake, H.; Hirakawa, H.; Shoji, M.; McBride, M.J.; Rhodes, R.G.; Nakayama, K. A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 276–281. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, T.; Wang, Y.; Muhammad, M.; Xue, W.; Ju, J.; Zhao, B. Knockout of alanine racemase gene attenuates the pathogenicity of Aeromonas hydrophila. BMC Microbiol. 2019, 19, 72. [Google Scholar] [CrossRef]
- Alvarez, B.; Secades, P.; McBride, M.J.; Guijarro, J.A. Development of genetic techniques for the psychrotrophic fish pathogen Flavobacterium psychrophilum. Appl. Environ. Microbiol. 2004, 70, 581–587. [Google Scholar] [CrossRef] [PubMed]

| Isolate | Origin | Isolation Year | Genome Size (Mb) | Genes | CDS | GC% | tRNA | Plasmid (kb) |
|---|---|---|---|---|---|---|---|---|
| FPS-G1 | Germany | 2017 | 2.86 | 2494 | 2423 | 32.4 | 49 | 3.3 |
| FPS-F15 | Finland | 2017 | 2.86 | 2496 | 2425 | 32.5 | 49 | 2.1 |
| FPS-P1 | Poland | 2016 | 2.86 | 2385 | 2344 | 32.5 | 41 | No |
| FPS-R9 | Russia | 2017 | 2.86 | 2387 | 2347 | 32.6 | 34 | No |
| FPS-P3 | Poland | 2017 | 2.86 | 2398 | 2358 | 32.4 | 34 | No |
| FPS-S6 | Sweden | 2017 | 2.86 | 2528 | 2457 | 32.5 | 49 | 3.3 |
| 160401-1/5N | Denmark | 2016 | 2.82 | 2527 | 2456 | 32.5 | 49 | 3.3 |
| P30-2B/09 | Finland | 2009 | 2.86 | 2542 | 2471 | 32.5 | 49 | 3.3 |
| F164 | Sweden | 1996 | 2.86 | 2489 | 2418 | 32.3 | 49 | No |
| FPS-F16 | Finland | 2017 | 2.86 | 2496 | 2425 | 32.5 | 49 | No |
| V46 | Finland | 2005 | 2.84 | 2294 | 2258 | 32.4 | 30 | No |
| FPS-S11A | Sweden | 2017 | 2.86 | 2484 | 2413 | 32.6 | 49 | 3.3 |
| 990512-1/2A | Denmark | 1999 | 2.86 | 2486 | 2415 | 32.5 | 49 | 3.3 |
| 141127-1/2N | Denmark | 2014 | 2.85 | 2488 | 2417 | 32.6 | 49 | 3.3 |
| P15-8B/11 | Finland | 2011 | 2.85 | 2489 | 2418 | 32.5 | 49 | 2.1 |
| 160401-1/5M | Denmark | 2016 | 2.85 | 2484 | 2413 | 32.5 | 49 | 3.3 |
| FPS-F27 | Finland | 2017 | 2.86 | 2490 | 2419 | 32.4 | 49 | 2.1 |
| FPS-R7 | Russia | 2017 | 3.20 | 2878 | 2806 | 32.6 | 49 | No |
| FPS-S9 | Sweden | 2017 | 2.86 | 2519 | 2448 | 32.5 | 49 | No |
| 030522-1/1 | Denmark | 2003 | 2.86 | 2534 | 2463 | 32.5 | 49 | 3.3 |
| FPS-S10 | Sweden | 2017 | 2.86 | 2.495 | 2425 | 32.5 | 49 | No |
| 010418-2/1 | Denmark | 2001 | 2.86 | 2546 | 2475 | 32.6 | 49 | 3.3 |
| K9/00 | Finland | 2000 | 2.86 | 2496 | 2425 | 32.5 | 49 | No |
| 950106-1/1 | Denmark | 1995 | 2.84 | 2507 | 2436 | 32.6 | 49 | 3.3 |
| NCIMB 1947T | USA | Unknown | 2.71 | 2397 | 2305 | 32.6 | 49 | No |
| FPS-S11B | Sweden | 2017 | 2.86 | 2486 | 2415 | 32.6 | 49 | 3.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castillo, D.; Donati, V.L.; Jørgensen, J.; Sundell, K.; Dalsgaard, I.; Madsen, L.; Wiklund, T.; Middelboe, M. Comparative Genomic Analyses of Flavobacterium psychrophilum Isolates Reveals New Putative Genetic Determinants of Virulence Traits. Microorganisms 2021, 9, 1658. https://doi.org/10.3390/microorganisms9081658
Castillo D, Donati VL, Jørgensen J, Sundell K, Dalsgaard I, Madsen L, Wiklund T, Middelboe M. Comparative Genomic Analyses of Flavobacterium psychrophilum Isolates Reveals New Putative Genetic Determinants of Virulence Traits. Microorganisms. 2021; 9(8):1658. https://doi.org/10.3390/microorganisms9081658
Chicago/Turabian StyleCastillo, Daniel, Valentina L. Donati, Jóhanna Jørgensen, Krister Sundell, Inger Dalsgaard, Lone Madsen, Tom Wiklund, and Mathias Middelboe. 2021. "Comparative Genomic Analyses of Flavobacterium psychrophilum Isolates Reveals New Putative Genetic Determinants of Virulence Traits" Microorganisms 9, no. 8: 1658. https://doi.org/10.3390/microorganisms9081658
APA StyleCastillo, D., Donati, V. L., Jørgensen, J., Sundell, K., Dalsgaard, I., Madsen, L., Wiklund, T., & Middelboe, M. (2021). Comparative Genomic Analyses of Flavobacterium psychrophilum Isolates Reveals New Putative Genetic Determinants of Virulence Traits. Microorganisms, 9(8), 1658. https://doi.org/10.3390/microorganisms9081658







