Endophytes from African Rice (Oryza glaberrima L.) Efficiently Colonize Asian Rice (Oryza sativa L.) Stimulating the Activity of Its Antioxidant Enzymes and Increasing the Content of Nitrogen, Carbon, and Chlorophyll
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of Endophytic Bacteria from O. glaberrima Rice Tissues
2.2. Bacterial Identification by 16S rRNA Sequence Analysis
2.2.1. DNA Extraction
2.2.2. PCR Amplification of the 16rRNA Gene Fragment
2.2.3. Cloning and Sequencing of the 16S Fragment
2.2.4. Phylogenetic Characterization
2.3. Genome Sequencing and Bioinformatic Analysis
2.4. Screening of Plant Growth-Promoting (PGP) Activities
2.4.1. Indole-3-Acetic Acid (IAA) Production
2.4.2. HPLC—Tandem Mass Spectrometry (HPLC-MS/MS)
2.5. PCR Amplification of the Nitrogenase Iron Protein (nifH) and ACC Deaminase (acdS) Genes Fragments
2.6. Plant Growth and Inoculation Methods
2.7. Re-Isolation of Diazotrophic Bacteria from Inoculated Plants
2.8. Strains Tagging with FPs
2.9. Confocal Laser Scanning Microscopy Detection
2.10. Nitrogenase Activity by Acetylene Reduction Assay (ARA)
2.10.1. Bacterial Cultures
2.10.2. Inoculated Plants
2.11. 15N2 Incorporation
2.12. Salt Stress
2.13. Antioxidant Enzymes Assays
2.14. Chlorophyll Content Measurement
2.15. Total Carbon and Nitrogen Analysis
2.16. Statistical Analysis
3. Results
3.1. Identification of Endophytic Bacteria
3.2. PGP-Traits of Bacterial Endophytes
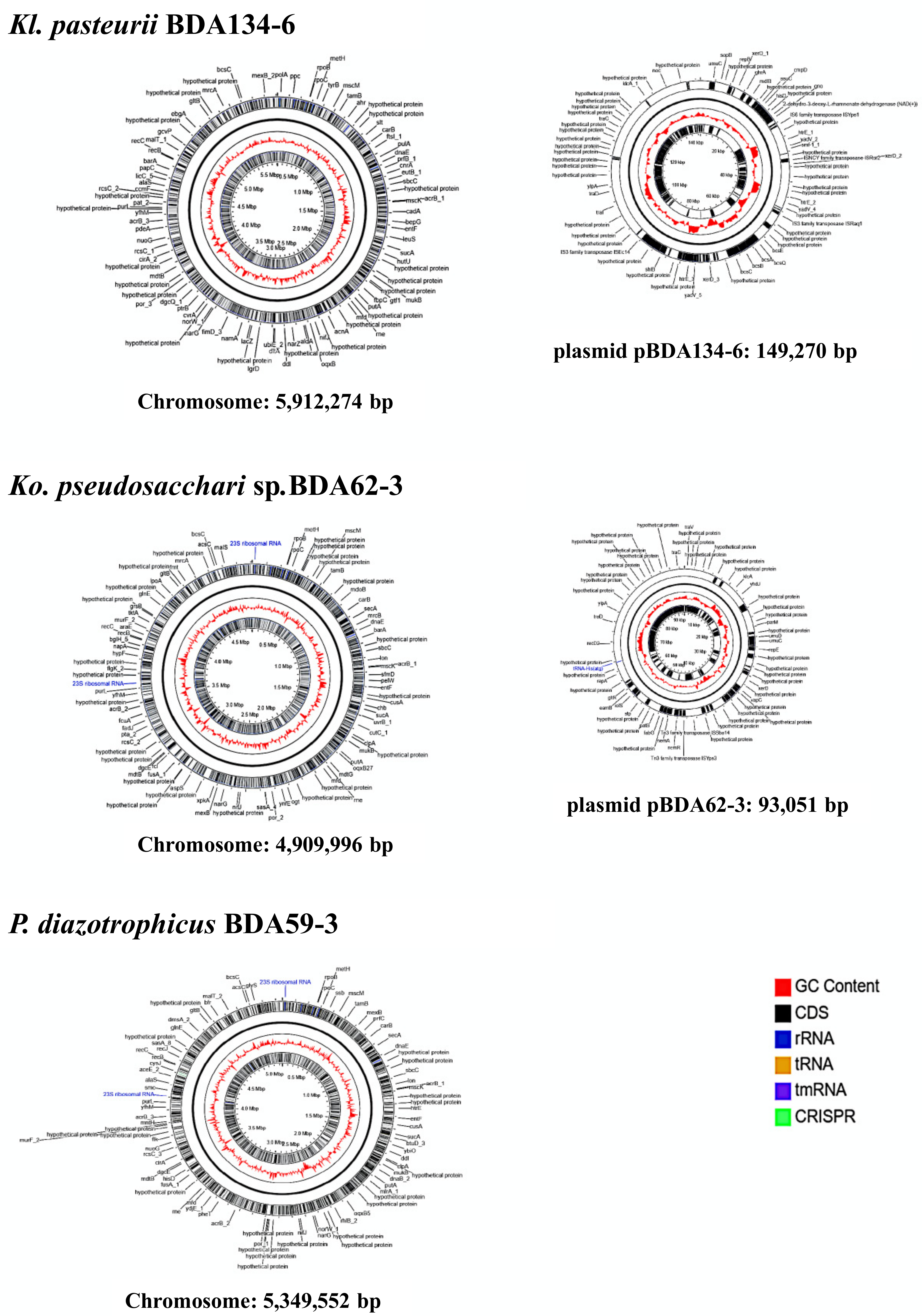
3.3. Genome Sequences of Kl. pasteurii BDA134-6, Ko. pseudosacchari BDA62-3 and P. diazotrophicus BDA59-3 Confirm the Presence of the Full Nitrogenase Genes Set
3.4. N-Fixing Endophytes Differently Colonize the Primary (O. glaberrima) and Secondary (O. sativa) Host
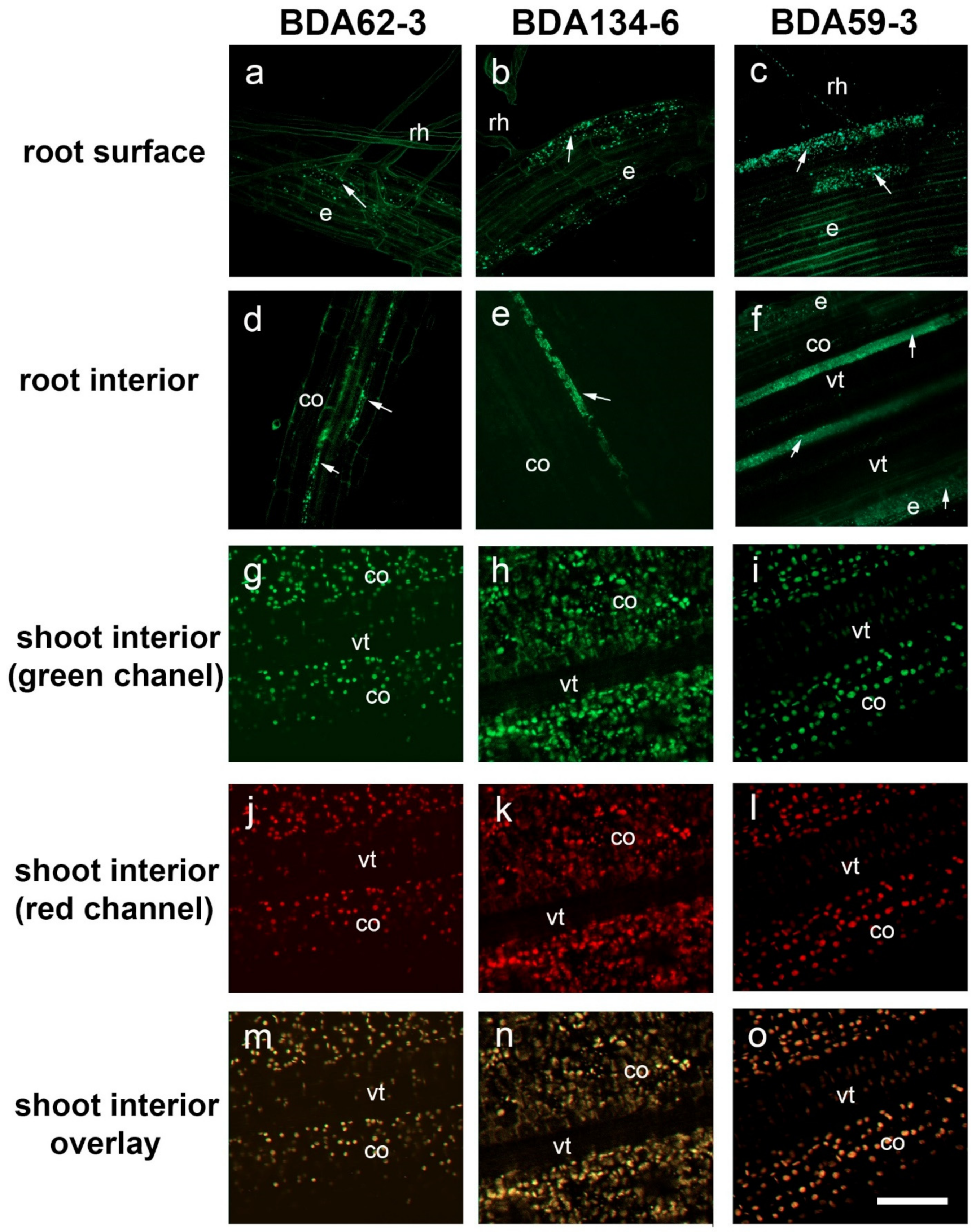
3.5. Colonization of Rice Roots by the Strains Ko. pseudosacchari BDA62-3, Kl. pasteurii BDA134-6 and P. diazotrophicus BDA59-3
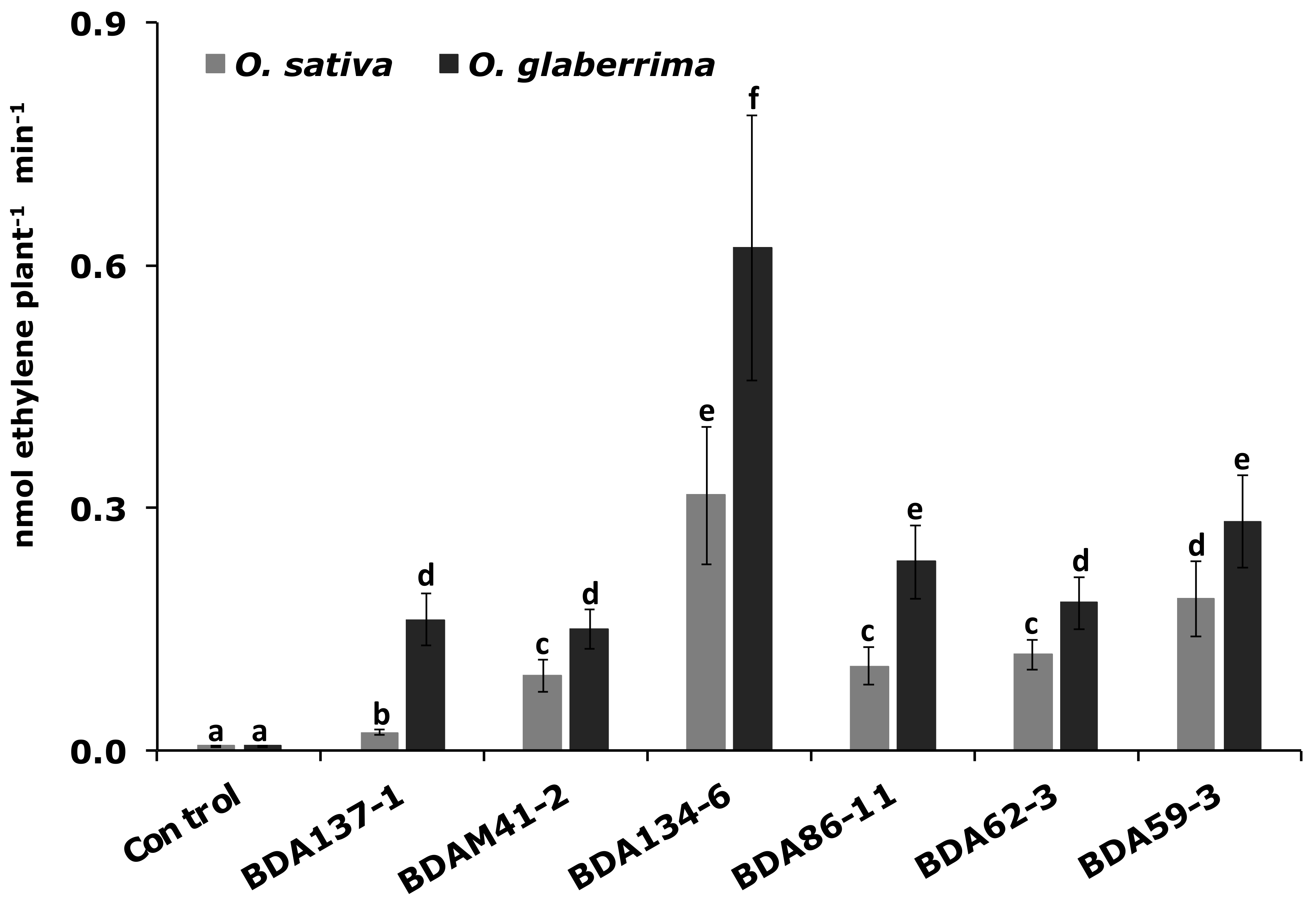
3.6. The Endophytes Kl. pasteurii BDA134-6 and P. diazotrophicus BDA59-3 Improve N-Fixation in Two Host Rice Species
3.7. N2 Gas Incorporation
3.8. Kl. pasteurii BDA134-6 and P. diazotrophicus BDA59-3 Increase Carbon (C), Nitrogen (N) and Chlorophyll (Chl) Content of Inoculated O. sativa Plants
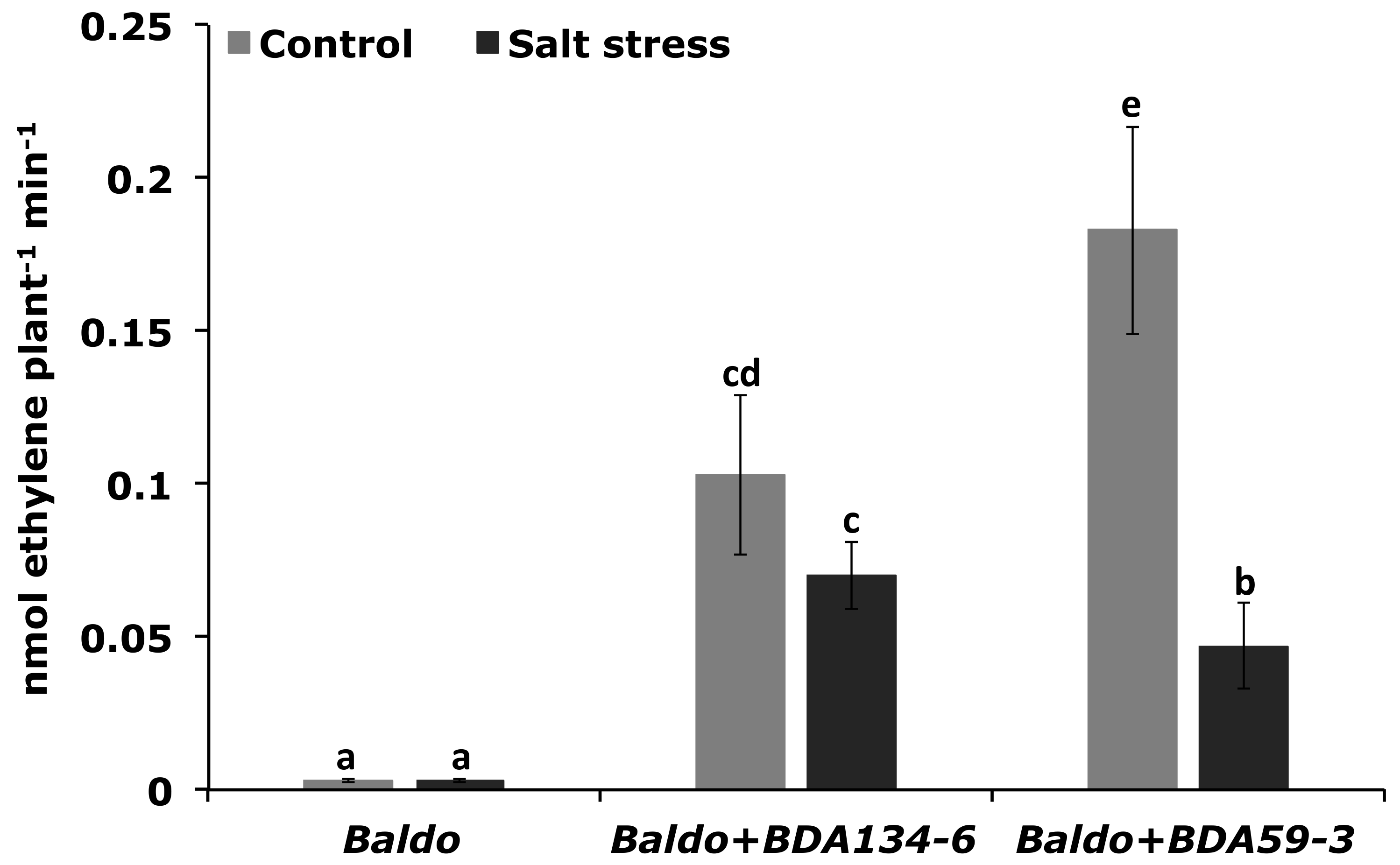
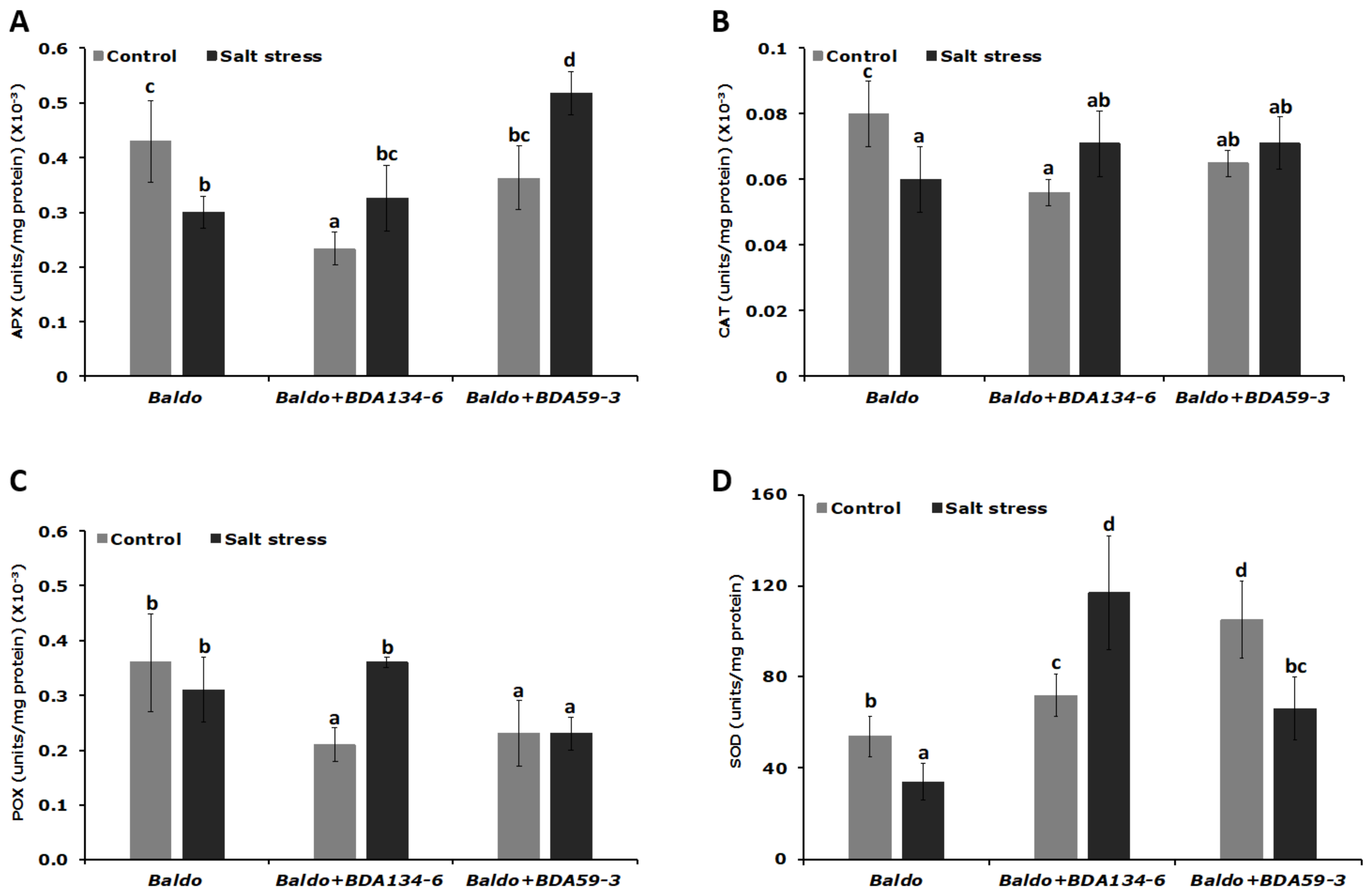
3.9. Inoculation of Rice Plants with Kl. pasteurii BDA134-6 Allows Maintenance of the Nitrogenase Function and Increases the Activity of Antioxidative Enzymes under Salinity Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kandel, S.L.; Joubert, P.M.; Doty, S.L. Bacterial Endophyte Colonization and Distribution within Plants. Microorganisms 2017, 5, 77. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Carvalhais, L.C.; Crawford, M.; Singh, E.; Dennis, P.G.; Pieterse, C.; Schenk, P.M. Inner Plant Values: Diversity, Colonization and Benefits from Endophytic Bacteria. Front. Microbiol. 2017, 8, 2552. [Google Scholar] [CrossRef] [PubMed]
- Olanrewaju, O.; Glick, B.R.; Babalola, O.O. Mechanisms of action of plant growth promoting bacteria. World J. Microbiol. Biotechnol. 2017, 33, 197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riyazuddin, R.; Verma, R.; Singh, K.; Nisha, N.; Keisham, M.; Bhati, K.K.; Kim, S.T.; Gupta, R. Ethylene: A Master Regulator of Salinity Stress Tolerance in Plants. Biomolecules 2020, 10, 959. [Google Scholar] [CrossRef]
- Gupta, B.; Huang, B. Mechanism of Salinity Tolerance in Plants: Physiological, Biochemical, and Molecular Characterization. Int. J. Genom. 2014, 2014, 701596. [Google Scholar] [CrossRef] [PubMed]
- Van Zelm, E.; Zhang, Y.; Testerink, C. Salt tolerance mechanisms of plants. Ann Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef] [Green Version]
- Sharma, P.; Jha, A.B.; Dubey, R.S.; Pessarakli, M. Reactive Oxygen Species, Oxidative Damage, and Antioxidative Defense Mechanism in Plants under Stressful Conditions. J. Bot. 2012, 2012, 217037. [Google Scholar] [CrossRef] [Green Version]
- Gupta, S.; Pandey, S. ACC Deaminase Producing Bacteria with Multifarious Plant Growth Promoting Traits Alleviates Salinity Stress in French Bean (Phaseolus vulgaris) Plants. Front. Microbiol. 2019, 10, 1506. [Google Scholar] [CrossRef]
- Kruasuwan, W.; Thamchaipenet, A. 1-Aminocyclopropane-1-carboxylate (ACC) Deaminase-Producing Endophytic Diazotrophic Enterobacter sp. EN-21 Modulates Salt–Stress Response in Sugarcane. J. Plant Growth Regul. 2018, 37, 849–858. [Google Scholar] [CrossRef]
- Tavares, M.J.; Nascimento, F.X.; Glick, B.R.; Rossi, M.J. The expression of an exogenous ACC deaminase by the endophyte Serratia grimesii BXF1 promotes the early nodulation and growth of common bean. Lett. Appl. Microbiol. 2018, 66, 252–259. [Google Scholar] [CrossRef]
- Linares, O.F. African rice (Oryza glaberrima): History and future potential. Proc. Natl. Acad. Sci. USA 2002, 99, 16360–16365. [Google Scholar] [CrossRef] [Green Version]
- Ndjiondjop, M.N.; Wambugu, P.; Sangare, J.R.; Dro, T.; Kpeki, B.; Gnikoua, K. Oryza glaberrima Steud. In The Wild Oryza Genomes. Compendium of Plant Genomes; Mondal, T., Henry, R., Eds.; Springer: Cham, Switzerland, 2018; pp. 105–126. [Google Scholar] [CrossRef]
- Van Andel, T. African Rice (Oryza glaberrima Steud.): Lost Crop of the Enslaved Africans Discovered in Suriname. Econ. Bot. 2010, 64, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Sie, M.; Sanni, K.; Futakuchi, K.; Manneh, B.; Mande, S.; Vodouhe, R.; Dogbe, S.; Dramé, K.-N.; Ogunbayo, A.; Ndjiondjop, M.-N.; et al. Towards a rational use of African rice (Oryza glaberrima Steud.) for breeding in Sub-Saharan Africa. Genes Genomes Genom. 2012, 6, 1–7. [Google Scholar]
- Andreozzi, A.; Prieto, P.; Mercado-Blanco, J.; Monaco, S.; Zampieri, E.; Romano, S.; Valè, G.; Defez, R.; Bianco, C. Efficient colonization of the endophytes Herbaspirillum huttiense RCA24 and Enterobacter cloacae RCA25 influences the physiological parameters of Oryza sativa L. cv. Baldo rice. Environ. Microbiol. 2018, 21, 3489–3504. [Google Scholar] [CrossRef]
- Edwards, J.; Johnson, C.; Santos-Medellin, C.; Lurie, E.; Podishetty, N.K.; Bhatnagar, S.; Eisen, J.A.; Sundaresan, V. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 2015, 112, E911–E920. [Google Scholar] [CrossRef] [Green Version]
- Santos-Medellin, C.; Edwards, J.; Liechty, Z.; Nguyen, B.; Sundaresan, V. Drought Stress Results in a Compartment-Specific Restructuring of the Rice Root-Associated Microbiomes. mBio 2017, 8, e00764-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Defez, R.; Andreozzi, A.; Bianco, C. The Overproduction of Indole-3-Acetic Acid (IAA) in Endophytes Upregulates Nitrogen Fixation in Both Bacterial Cultures and Inoculated Rice Plants. Microb. Ecol. 2017, 74, 441–452. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Ardui, S.; Ameur, A.; Vermeesch, J.R.; Hestand, M.S. Single molecule real-time (SMRT) sequencing comes of age: Applications and utilities for medical diagnostics. Nucleic Acids Res. 2018, 46, 2159–2168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Cuccuru, G.; Orsini, M.; Pinna, A.; Sbardellati, A.; Soranzo, N.; Travaglione, A.; Uva, P.; Zanetti, G.; Fotia, G. Orione, a web-based framework for NGS analysis in microbiology. Bioinformatics 2014, 30, 1928–1929. [Google Scholar] [CrossRef] [Green Version]
- Lee, I.; Chalita, M.; Ha, S.-M.; Na, S.-I.; Yoon, S.-H.; Chun, J. ContEst16S: An algorithm that identifies contaminated prokaryotic genomes using 16S RNA gene sequences. Int. J. Syst. Evol. Microbiol. 2017, 67, 2053–2057. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Grissa, I.; Vergnaud, G.; Pourcel, C. CRISPRFinder: A web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007, 35, W52–W57. [Google Scholar] [CrossRef] [Green Version]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [Green Version]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. KAAS: An automatis genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007, 35, W182–W185. [Google Scholar] [CrossRef] [Green Version]
- Hui, X.; Chen, Z.; Lin, M.; Zhang, J.; Hu, Y.; Zeng, Y.; Cheng, X.; Ou-Yang, L.; Sun, M.-A.; White, A.P.; et al. T3SEpp: An Integrated Prediction Pipeline for Bacterial Type III Secreted Effectors. mSystems 2020, 5, e00288-20. [Google Scholar] [CrossRef] [PubMed]
- Salkowski, E. Uber das verhalten der skatolcarbonsaure im organismus. Z. Physiol. Chem. 1885, 9, 23–33. [Google Scholar]
- Gaby, J.; Buckley, D.H. A Comprehensive Evaluation of PCR Primers to Amplify the nifH Gene of Nitrogenase. PLoS ONE 2012, 7, e42149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Chang, S.; Ye, S.; Chen, M.; Lin, L.; Li, Y.; Li, S.; An, Q. Differentiation of 1-aminocyclopropane-1-carboxylate (ACC) deaminase from its homologs is the key for identifying bacteria containing ACC deaminase. FEMS Microbiol. Ecol. 2015, 91, fiv112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, S.; Xiao, Y.; Xu, F.; Gao, X.; Cao, S.; Zhang, F.; Wang, G.; Sanders, D.; Chu, C. Cytokinin-dependent regulatory module underlies the maintenance of zinc nutrition in rice. New Phytol. 2019, 224, 202–215. [Google Scholar] [CrossRef] [PubMed]
- Bloemberg, G.V.; Wijfjes, A.H.M.; Lamers, G.E.M.; Stuurman, N.; Lugtenberg, B.J.J. Simultaneous Imaging of Pseudomonas fluorescens WCS365 Populations Expressing Three Different Autofluorescent Proteins in the Rhizosphere: New Perspectives for Studying Microbial Communities. Mol. Plant Microbe Interact. 2000, 13, 1170–1176. [Google Scholar] [CrossRef] [Green Version]
- Prieto, P.; Mercado-Blanco, J. Endophytic colonization of olive roots by the biocontrol strain Pseudomonas fluorescens PICF7. FEMS Microbiol. Ecol. 2008, 64, 297–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Defez, R.; Esposito, R.; Angelini, C.; Bianco, C. Overproduction of Indole-3-Acetic Acid in Free-Living Rhizobia Induces Transcriptional Changes Resembling Those Occurring in Nodule Bacteroids. Mol. Plant Microbe Interact. 2016, 29, 484–495. [Google Scholar] [CrossRef] [Green Version]
- Stiefel, E.I. The mechanism of nitrogen fixation. In Recent Development of Nitrogen Fixation; Newton, W., Postgate, J.R., Rodriguez-Barrueco, C., Eds.; Academic Press: London, UK, 1977; pp. 69–108. [Google Scholar]
- Brand, W.A.; Coplen, T.B. Stable isotope deltas: Tiny, yet robust signatures in nature. Isot. Environ. Health Stud. 2012, 48, 393–409. [Google Scholar] [CrossRef] [PubMed]
- Bianco, C.; Defez, R. Medicago truncatula improves salt tolerance when nodulated by an indole-3-acetic acid-overproducing Sinorhizobium meliloti strain. J. Exp. Bot. 2009, 60, 3097–3107. [Google Scholar] [CrossRef]
- Liang, Y.; Urano, D.; Liao, K.-L.; Hedrick, T.L.; Gao, Y.; Jones, A.M. A nondestructive method to estimate the chlorophyll content of Arabidopsis seedlings. Plant Methods 2017, 13, 26. [Google Scholar] [CrossRef]
- Arnon, D.I. Copper enzymes in isolated chloroplasts. polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949, 24, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.X.; Peng, G.X.; Wang, E.T.; Yan, H.; Yuan, Q.H.; Zhang, W.; Lou, X.; Wu, H.; Tan, Z.Y. Diverse endophytic nitrogen-fixing bacteria isolated from wild rice Oryza rufipogon and description of Phytobacter diazotrophicus gen. nov. sp. nov. Arch. Microbiol. 2007, 189, 431–439. [Google Scholar] [CrossRef]
- Pillonetto, M.; Arend, L.N.; Faoro, H.; D’Espindula, H.; Blom, J.; Smits, T.H.; Mira, M.; Rezzonico, F. Emended description of the genus Phytobacter, its type species Phytobacter diazotrophicus (Zhang 2008) and description of Phytobacter ursingii sp. nov. Int. J. Syst. Evol. Microbiol. 2018, 68, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Borenshtein, D.; Schauer, D.B. The genus Citrobacter. In The Prokaryotes; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006. [Google Scholar] [CrossRef]
- Bakker, A.; Smith, D.W. Methylation of GATC sites is required for precise timing between rounds of DNA replication in Escherichia coli. J. Bacteriol. 1989, 171, 5738–5742. [Google Scholar] [CrossRef] [Green Version]
- Levy, A.; Gonzalez, I.S.; Mittelviefhaus, M.; Clingenpeel, S.; Paredes, S.H.; Miao, J.; Wang, K.; Devescovi, G.; Stillman, K.; Monteiro, F.; et al. Genomic features of bacterial adaptation to plants. Nat. Genet. 2017, 50, 138–150. [Google Scholar] [CrossRef] [Green Version]
- Han, A.W.; Sandy, M.; Fishman, B.; Trindade-Silva, A.E.; Soares, C.A.G.; Distel, D.L.; Butler, A.; Haygood, M.G. Turnerbactin, a Novel Triscatecholate Siderophore from the Shipworm Endosymbiont Teredinibacter turnerae T7901. PLoS ONE 2013, 8, e76151. [Google Scholar] [CrossRef] [Green Version]
- Spraker, J.E.; Sanchez, L.M.; Lowe, T.M.; Dorrestein, P.C.; Keller, N.P. Ralstonia solanacearum lipopeptide induces chlmydospore development in fungi and facilitates bacterial entry into fungal tissues. ISME J. 2016, 10, 1317–2330. [Google Scholar] [CrossRef] [Green Version]
- Unterhauser, K.; Pöltl, L.; Schneditz, G.; Kienesberger, S.; Glabonjat, R.; Kitsera, M.; Pletz, J.; Prado, F.J.; Dornisch, E.; Lembacher-Fadum, C.; et al. Klebsiella oxytoca enterotoxins tilimycin and tilivalline have distinct host DNA-damaging and microtubule-stabilizing activities. Proc. Natl. Acad. Sci. USA 2019, 116, 3774–3783. [Google Scholar] [CrossRef] [Green Version]
- Shibu, P.; McCuaig, F.; McCartney, A.L.; Kujawska, M.; Hall, L.J.; Hoyles, L. Improved molecular characterization of the the Klebsiella oxytoca complex reveals the presence of the kleboxymycin biosynthesis gene cluster. bioRxiv 2020. [Google Scholar] [CrossRef]
- Pérez-Jaramillo, J.E.; Mendes, R.; Raaijmakers, J.M. Impact of plant domestication on rhizosphere microbiome assembly and functions. Plant Mol. Biol. 2015, 90, 635–644. [Google Scholar] [CrossRef] [Green Version]
- Correa-Galeote, D.; Bedmar, E.J.; Arone, G. Maize Endophytic Bacterial Diversity as Affected by Soil Cultivation History. Front. Microbiol. 2018, 9, 484. [Google Scholar] [CrossRef] [PubMed]
- Etminani, F.; Harighi, B. Isolation and Identification of Endophytic Bacteria with Plant Growth Promoting Activity and Biocontrol Potential from wild Pistachio Trees. Plant Pathol. J. 2018, 34, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Brígido, C.; Singh, S.; Menéndez, E.; Tavares, M.J.; Glick, B.R.; Félix, M.D.R.; Oliveira, S.; Carvalho, M. Diversity and Functionality of Culturable Endophytic Bacterial Communities in Chickpea Plants. Plants 2019, 8, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertani, I.; Abbruscato, P.; Piffanelli, P.; Subramoni, S.; Venturi, V. Rice bacterial endophytes: Isolation of a collection, identification of beneficial strains and microbiome analysis. Environ. Microbiol. Rep. 2016, 8, 388–398. [Google Scholar] [CrossRef]
- Moronta-Barrios, F.; Gionechetti, F.; Pallavicini, A.; Marys, E.; Venturi, V. Bacterial Microbiota of Rice Roots: 16S-Based Taxonomic Profiling of Endophytic and Rhizospheric Diversity, Endophytes Isolation and Simplified Endophytic Community. Microorganisms 2018, 6, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greetatorn, T.; Hashimoto, S.; Sarapat, S.; Tittabutr, P.; Boonkerd, N.; Uchiumi, T.; Teaumroong, N. Empowering rice seedling growth by endophytic Bradyrhizobium sp. SUTN 9-2. Lett. Appl. Microbiol. 2019, 68, 258–266. [Google Scholar] [CrossRef]
- Afzal, I.; Shinwari, Z.K.; Sikandar, S.; Shahzad, S. Plant beneficial endophytic bacteria: Mechanisms, diversity, host range and genetic determinants. Microbiol. Res. 2019, 221, 36–49. [Google Scholar] [CrossRef]
- Ljung, K. Auxin metabolism and homeostasis during plant development. Development 2013, 140, 943–950. [Google Scholar] [CrossRef] [Green Version]
- Olatunji, D.; Geelen, D.; Verstraeten, I. Control of Endogenous Auxin Levels in Plant Root Development. Int. J. Mol. Sci. 2017, 18, 2587. [Google Scholar] [CrossRef] [Green Version]
- Spaepen, S.; Vanderleyden, J.; Remans, R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 2007, 31, 425–448. [Google Scholar] [CrossRef] [Green Version]
- Egamberdieva, D.; Wirth, S.; Alqarawi, A.; Abd Allah, E.; Hashem, A. Phytohormones and Beneficial Microbes: Essential Components for Plants to Balance Stress and Fitness. Front. Microbiol. 2017, 8, 2104. [Google Scholar] [CrossRef] [PubMed]
- Hill, S. Physiology of nitrogen fixation in free-living heterotrophs. In Biological Nitrogen Fixation; Stacey, G., Burris, R.H., Evans, H.J., Eds.; Chapman Hall: New York, NY, USA, 1992; pp. 87–134. [Google Scholar]
- Singha, K.M.; Singh, B.; Pandey, P. Host specific endophytic microbiome diversity and associated functions in three varieties of scented black rice are dependent on growth stage. Sci. Rep. 2021, 11, 12259. [Google Scholar] [CrossRef] [PubMed]
- Ndjiondjop, M.N.; Cisse, F.; Girma, G.; Sow, M.; Bocco, R.; Djedatin, G.; Blandine, F. Morpho-agronomic and molecular characterisation of Oryza glaberrima germplasm from Mali. Afr. J. Biotechnol. 2010, 9, 7409–7417. [Google Scholar] [CrossRef]
- Wambugu, P.W.; Ndjiondjop, M.-N.; Henry, R. Advances in Molecular Genetics and Genomics of African Rice (Oryza glaberrima Steud). Plants 2019, 8, 376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mosquito, S.; Bertani, I.; Licastro, D.; Compant, S.; Myers, M.P.; Hinarejos, E.; Levy, A.; Venturi, V. In Planta Colonization and Role of T6SS in Two Rice Kosakonia Endophytes. Mol. Plant Microbe Interact. 2020, 33, 349–363. [Google Scholar] [CrossRef] [PubMed]
- Udwardi, M.; Poole, P.S. Transport and metabolism in legume-rhizobia symbioses. Ann. Rev. Plant Biol. 2013, 64, 781–805. [Google Scholar] [CrossRef] [Green Version]
- Merla, C.; Rodrigues, C.; Passet, V.; Corbella, M.; Thorpe, H.A.; Kallonen, T.V.S.; Zong, Z.; Marone, P.; Bandi, C.; Sassera, D.; et al. Description of Klebsiella spallanzanii sp. nov. and of Klebsiella pasteurii sp. nov. Front. Microbiol. 2019, 10, 2360. [Google Scholar] [CrossRef] [Green Version]
- Bernal, P.; Llamas, M.A.; Filloux, A. Type VI secretion systems in plant-associated bacteria. Environ. Microbiol. 2017, 20, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Defez, R.; Andreozzi, A.; Romano, S.; Pocsfalvi, G.; Fiume, I.; Esposito, R.; Angelini, C.; Bianco, C. Bacterial IAA-Delivery into Medicago Root Nodules Triggers a Balanced Stimulation of C and N Metabolism Leading to a Biomass Increase. Microorganisms 2019, 7, 403. [Google Scholar] [CrossRef] [Green Version]


| Strain | Nitrogenase Activity § (nmol Ethylene/mg Cells/min) | IAA (nmol/mg Cells) ξ |
|---|---|---|
| Ko. oryzendophytica BDA137-1 | 0.032 ± 0.004 c | 8.7 ± 1.4 c |
| Enterobacter sp. BDAM41-2 | 0.025 ± 0.005 c | 6.1 ± 0.3 c |
| Kl. pasteurii BDA134-6 | 0.160 ± 0.024 a | 31 ± 2 b |
| E. sacchari BDA86-11 | 0.064 ± 0.008 b | 7.4 ± 0.3 c |
| Ko pseudosacchari BDA62-3 | 0.049 ± 0.008 b | 7.7 ± 0.5 c |
| P. diazotrophicus BDA59-3 | 0.044 ± 0.003 b | 61 ± 10 a |
| Gene Code | Gene Name |
|---|---|
| Kl. pasteurii BDA134-6 | |
| BDA134-6_02421 | nifJ |
| BDA134-6_03551 | nifH |
| BDA134-6_03552 | nifD |
| BDA134-6_03553 | nifK_1 |
| BDA134-6_03556 | nifD_2 |
| BDA134-6_03557 | nifK_2 |
| BDA134-6_03560 | nifS |
| BDA134-6_03562 | nifW |
| BDA134-6_03565 | nifF |
| BDA134-6_03566 | nifL |
| BDA134-6_03567 | nifA |
| BDA134-6_03568 | nifB |
| Ko. pseudosacchari BDA62-3 | |
| Entero_BDA62.3_02197 | nifQ |
| Entero_BDA62.3_02198 | nifB |
| Entero_BDA62.3_02199 | nifA |
| Entero_BDA62.3_02200 | nifL |
| Entero_BDA62.3_02201 | nifF |
| Entero_BDA62.3_02203 | nifZ |
| Entero_BDA62.3_02204 | nifW |
| Entero_BDA62.3_02206 | nifS |
| Entero_BDA62.3_02207 | nifU |
| Entero_BDA62.3_02208 | Dinitrogenase iron-molybdenum cofactor |
| Entero_BDA62.3_02209 | nifK_1 |
| Entero_BDA62.3_02210 | nifD_1 |
| Entero_BDA62.3_02211 | Dinitrogenase iron-molybdenum cofactor |
| Entero_BDA62.3_02212 | nifT/fixU |
| Entero_BDA62.3_02213 | niK_2 |
| Entero_BDA62.3_02214 | nifD_2 |
| Entero_BDA62.3_02215 | nifH |
| P. diazotrophicus BDA59-3 | |
| Citro_BDA59-3_02396 | nifJ_1 |
| Citro_BDA59-3_02623 | nifJ_2 |
| Citro_BDA59-3_02624 | nifH |
| Citro_BDA59-3_02625 | nifD_1 |
| Citro_BDA59-3_02626 | nifK_1 |
| Citro_BDA59-3_02627 | nifT/fixU |
| Citro_BDA59-3_02628 | Dinitrogenase iron-molybdenum cofactor |
| Citro_BDA59-3_02629 | nifD_2 |
| Citro_BDA59-3_02630 | nifK_2 |
| Citro_BDA59-3_02631 | Dinitrogenase iron-molybdenum cofactor |
| Citro_BDA59-3_02632 | nifU |
| Citro_BDA59-3_02633 | nifS |
| Citro_BDA59-3_02635 | nifW |
| Citro_BDA59-3_02636 | nifZ |
| Citro_BDA59-3_02638 | nifF |
| Citro_BDA59-3_02639 | nifL |
| Citro_BDA59-3_02640 | nifA |
| Citro_BDA59-3_02641 | nifB |
| Strain | O. glaberrima (CFU per Plant) § (×10 5) | O. sativa (CFU per Plant) § (×10 5) |
|---|---|---|
| Ko. oryzendophytica BDA137-1 | 151 ± 24 e | 10 ± 2 d |
| Enterobacter sp. BDAM41-2 | 2264 ± 410 a | 10 ± 2 d |
| Kl. pasteuri BDA134-6 | 1064 ± 93 b | 176 ± 39 a |
| E. sacchari BDA86-11 | 613 ± 76 c | 180 ± 38 a |
| Ko. pseudosacchari BDA62-3 | 312 ± 92 d | 154 ± 24 b |
| P. diazotrophicus BDA59-3 | 384 ± 57 d | 49 ± 8 c |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bianco, C.; Andreozzi, A.; Romano, S.; Fagorzi, C.; Cangioli, L.; Prieto, P.; Cisse, F.; Niangado, O.; Sidibé, A.; Pianezze, S.; et al. Endophytes from African Rice (Oryza glaberrima L.) Efficiently Colonize Asian Rice (Oryza sativa L.) Stimulating the Activity of Its Antioxidant Enzymes and Increasing the Content of Nitrogen, Carbon, and Chlorophyll. Microorganisms 2021, 9, 1714. https://doi.org/10.3390/microorganisms9081714
Bianco C, Andreozzi A, Romano S, Fagorzi C, Cangioli L, Prieto P, Cisse F, Niangado O, Sidibé A, Pianezze S, et al. Endophytes from African Rice (Oryza glaberrima L.) Efficiently Colonize Asian Rice (Oryza sativa L.) Stimulating the Activity of Its Antioxidant Enzymes and Increasing the Content of Nitrogen, Carbon, and Chlorophyll. Microorganisms. 2021; 9(8):1714. https://doi.org/10.3390/microorganisms9081714
Chicago/Turabian StyleBianco, Carmen, Anna Andreozzi, Silvia Romano, Camilla Fagorzi, Lisa Cangioli, Pilar Prieto, Fousseyni Cisse, Oumar Niangado, Amadou Sidibé, Silvia Pianezze, and et al. 2021. "Endophytes from African Rice (Oryza glaberrima L.) Efficiently Colonize Asian Rice (Oryza sativa L.) Stimulating the Activity of Its Antioxidant Enzymes and Increasing the Content of Nitrogen, Carbon, and Chlorophyll" Microorganisms 9, no. 8: 1714. https://doi.org/10.3390/microorganisms9081714
APA StyleBianco, C., Andreozzi, A., Romano, S., Fagorzi, C., Cangioli, L., Prieto, P., Cisse, F., Niangado, O., Sidibé, A., Pianezze, S., Perini, M., Mengoni, A., & Defez, R. (2021). Endophytes from African Rice (Oryza glaberrima L.) Efficiently Colonize Asian Rice (Oryza sativa L.) Stimulating the Activity of Its Antioxidant Enzymes and Increasing the Content of Nitrogen, Carbon, and Chlorophyll. Microorganisms, 9(8), 1714. https://doi.org/10.3390/microorganisms9081714











