Depth-Dependent Spatiotemporal Dynamics of Overwintering Pelagic Microcystis in a Temperate Water Body
Abstract
1. Introduction
2. Materials and Methods
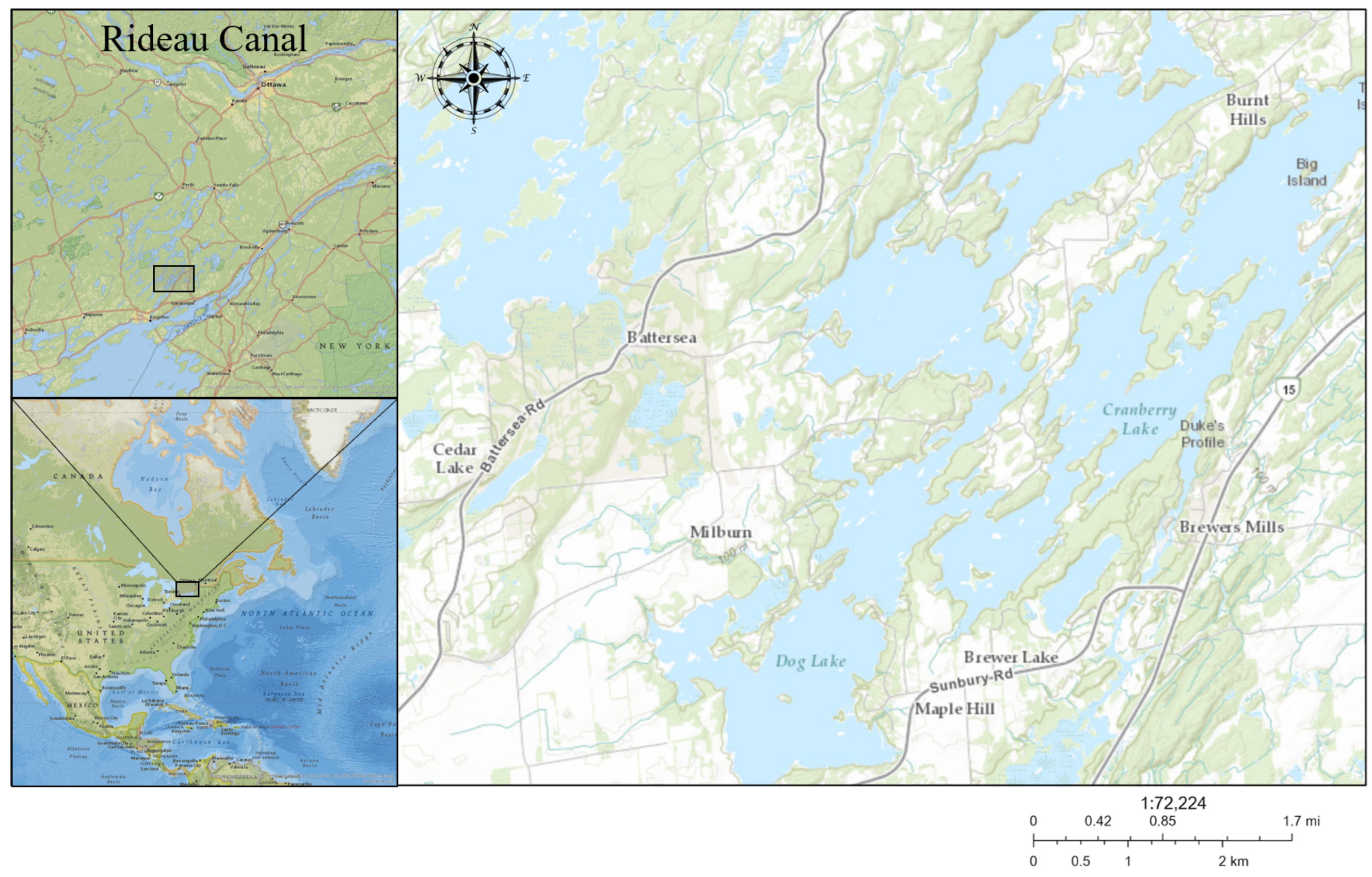
2.1. Study Site
2.2. Sampling Scheme
2.3. Sample Filtration
2.4. DNA Extraction
2.5. Primer Design and Standards
2.6. Quantitative PCR
2.7. Statistics
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kiørboe, T.; Hansen, J.; Alldredge, A.; Jackson, G.; Passow, U.; Dam, H.; Drapeau, D.; Waite, A.; Garcia, C. Sedimentation of phytoplankton during a diatom bloom: Rates and mechanisms. J. Mar. Res. 1996, 54, 1123–1148. [Google Scholar] [CrossRef]
- Richardson, K. Harmful or exceptional phytoplankton blooms in the marine ecosystem. Adv. Mar. Biol. 1997, 31, 301–385. [Google Scholar] [CrossRef]
- Havens, K. Cyanobacteria blooms: Effects on aquatic ecosystems. Adv. Exp. Med. Biol. 2008, 619, 733–747. [Google Scholar] [CrossRef]
- Funkey, C.P.; Conley, D.J.; Reuss, N.S.; Humborg, C.; Jilbert, T.; Slomp, C.P. Hypoxia sustains cyanobacteria blooms in the Baltic Sea. Environ. Sci. Technol. 2014, 48, 2598–2602. [Google Scholar] [CrossRef]
- Filstrup, C.; Hillebrand, H.; Heathcote, A.; Harpole, W.; Downing, J. Cyanobacteria dominance influences resource use efficiency and community turnover in phytoplankton and zooplankton communities. Ecol. Lett. 2014, 17, 464–474. [Google Scholar] [CrossRef]
- Carmichael, W.W.; Azevedo, S.M.; An, J.S.; Molica, R.J.; Jochimsen, E.M.; Lau, S.; Rinehart, K.L.; Shaw, G.R.; Eaglesham, G.K. Human fatalities from cyanobacteria: Chemical and biological evidence for cyanotoxins. Environ. Health Perspect. 2001, 109, 663–668. [Google Scholar] [CrossRef] [PubMed]
- Puschner, B.; Hoff, B.; Tor, E.R. Diagnosis of anatoxin-a poisoning in dogs from North America. J. Vet. Diagn. Investig. 2008, 20, 89–92. [Google Scholar] [CrossRef]
- Mez, K.; Beattie, K.A.; Codd, G.A.; Hanselmann, K.; Hauser, B.; Naegeli, H.; Preisig, H.R. Identification of a microcystin in benthic cyanobacteria linked to cattle deaths on alpine pastures in Switzerland. Eur. J. Phycol. 1997, 32, 111–117. [Google Scholar] [CrossRef]
- Downing, J.; Watson, S.; McCauley, E. Predicting cyanobacteria dominance in lakes. Can. J. Fish. Aquat. Sci. 2001, 58, 1905–1908. [Google Scholar] [CrossRef]
- Paerl, H.W.; Huisman, J. Climate change: A catalyst for global expansion of harmful cyanobacterial blooms. Environ. Microbiol. Rep. 2009, 1, 27–37. [Google Scholar] [CrossRef]
- Paerl, H.W.; Ustach, J.F. Blue-green algal scums: An explanation for their occurrence during freshwater blooms. Limnol. Oceanogr. 1982, 27, 212–217. [Google Scholar] [CrossRef]
- De Senerpont Domis, L.; Mooij, W.; Huisman, J. Climate-induced shifts in an experimental phytoplankton community: A mechanistic approach. Hydrobiologia 2007, 584, 403–413. [Google Scholar] [CrossRef]
- Jankowaik, J.; Hattenrath-Lehmann, T.; Kramer, B.J.; Ladds, M.; Gobbler, C.J. Deciphering the effects of nitrogen, phosphorus, and temperature on cyanobacterial bloom intensity, diversity, and toxicity in western Lake Erie. Limnol. Oceanogr. 2019, 64, 1347–1370. [Google Scholar] [CrossRef]
- Smucker, N.J.; Beaulieu, J.J.; Nietch, C.T.; Young, J.L. Increasingly severe cyanobacterial blooms and deep water hypoxia coincide with warming water temperatures in reservoirs. Glob. Chang. Biol. 2021, 21, 2507–2519. [Google Scholar] [CrossRef] [PubMed]
- Kosten, S.; Huszar, V.L.M.; Bécares, E.; Costa, L.S.; van Donk, E.; Hansson, L.-A.; Jeppesen, E.; Kruk, C.; Lacerot, G.; Mazzeo, N.; et al. Warmer climates boost cyanobacterial dominance in shallow lakes. Glob. Chang. Biol. 2011, 18, 118–126. [Google Scholar] [CrossRef]
- Paerl, H.W.; Huisman, J. Blooms like it hot. Science 2008, 320, 57–58. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, C.; Ding, J.; Hou, A.; Li, Y.; Niu, Z.; Su, X.; Xu, Y.; Laws, E.A. The 2007 water crisis in Wuxi, China: Analysis of the origin. J. Hazard. Mater. 2010, 182, 130–135. [Google Scholar] [CrossRef]
- Michalak, A.M.; Anderson, E.J.; Beletsky, D.; Boland, S.; Bosch, N.S.; Bridgeman, T.B.; Chaffin, J.D.; Cho, K.; Confesor, R.; Daloglu, I.; et al. Record-setting algal bloom in Lake Erie caused by agricultural and meteorological trends consistent with expected future conditions. Proc. Natl. Acad. Sci. USA 2013, 110, 6448–6452. [Google Scholar] [CrossRef] [PubMed]
- Orihel, D.M.; Baulch, H.M.; Casson, N.J.; North, R.L.; Parsons, C.T.; Seckar, D.C.M.; Venkiteswaran, J.J. Internal phosphorus loading in Canadian fresh waters: A critical review and data analysis. Can. J. Fish. Aquat. Sci. 2017, 74, 2005–2029. [Google Scholar] [CrossRef]
- Valdemarsen, T.B.; Quintana, C.O.; Flindt, M.R.; Kristensen, E. Organic N and P in eutrophic fjord sediments—Rates of mineralization and consequences for internal nutrient loading. Biogeosciences 2015, 12, 1765–1779. [Google Scholar] [CrossRef]
- Qu, M.; Lefebvre, D.D.; Wang, Y.; Qu, Y.; Zhu, D.; Ren, W. Algal blooms: Proactive strategy. Science 2014, 10, 175–176. [Google Scholar] [CrossRef]
- Visser, P.M.; Ibelings, B.W.; Mur, L.R.; Walsby, A.E. The ecophysiology of the harmful cyanobacterium Microcystis. In Harmful Cyanobacteria; Huisman, J., Matthijs, H.C., Visser, P.M., Eds.; Springer: Dordrecht, Germany, 2005; pp. 109–142. [Google Scholar]
- Walsby, A.E. Gas vesicles. Microbiol. Rev. 1994, 58, 94–144. [Google Scholar] [CrossRef] [PubMed]
- Thomas, R.H.; Walsby, A.E. Buoyancy regulation in a strain of Microcystis. Microbiology 1985, 131, 799–809. [Google Scholar] [CrossRef]
- Visser, P.M.; Ibelings, B.W.; Mur, L.R. Autumnal sedimentation of Microcystis spp. as result of an increase in carbohydrate ballast at reduced temperature. J. Plankton Res. 1995, 17, 919–933. [Google Scholar] [CrossRef]
- Vinh, L.; Tanabe, Y.; Matsuura, H.; Kaya, K.; Watanabe, M. Morphological, biochemical and phylogenetic assessments of water-bloom-forming tropical morphospecies of Microcystis. Phycol. Res. 2012, 60, 208–222. [Google Scholar] [CrossRef]
- Brunberg, A.-K.; Blomqvist, P. Benthic overwintering of Microcystis colonies under different environmental conditions. J. Plankton Res. 2002, 24, 1247–1252. [Google Scholar] [CrossRef]
- Reynolds, C.S.; Jaworsky, G.H.M.; Cmiech, H.A.; Leedale, G.F. On the annual cycle of the blue-green algae Microcystis aeruginosa Kütz. emend. Elenkin. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1981, 293, 419–477. [Google Scholar] [CrossRef]
- Verspagen, J.M.H.; Snelder, E.; Visser, P.M.; Jöhnk, K.D.; Ibelings, B.W.; Mur, L.R.; Huisman, J. Benthic-pelagic coupling in the population dynamics of the harmful cyanobacterium Microcystis. Freshw. Biol. 2005, 50, 854–867. [Google Scholar] [CrossRef]
- Verspagen, J.M.H.; Snelder, E.; Visser, P.M.; Huisman, J.; Mur, L.R.; Ibelings, B.W. Recruitment of benthic Microcystis (Cyanophyceae) to the water column: Internal buoyancy changes or resuspension? J. Phycol. 2004, 40, 260–270. [Google Scholar] [CrossRef]
- Bade, D.L. Lake ecosystems (Stratification and seasonal mixing processes, pelagic and benthic coupling). In Encyclopedia of Hydrological Sciences; Anderson, M.G., McDonnell, J.J., Eds.; Wiley: Hoboken, NJ, USA, 2006. [Google Scholar]
- Vasas, G.; Baecsi, I.; Suranyi, G.; Hamvas, M.M.; Mathe, C.; Nagy, S.A.; Borbely, G. Isolation of viable cell mass from frozen Microcystis viridis bloom containing microcystin-RR. Hydrobiologia 2010, 639, 147–151. [Google Scholar] [CrossRef]
- Núñez-Vázquez, E.J.; Tovar-Ramírez, D.; Heredia-Tapia, A.; Ochoa, J. Freeze survival of the cyanobacteria Microcoleus chthonoplastes without cryoprotector. J. Environ. Biol. 2011, 32, 407–417. [Google Scholar] [PubMed]
- Park, H.-K. Long-term preservation of bloom-forming cyanobacteria by cryopreservation. Algae 2006, 21, 125–131. [Google Scholar] [CrossRef]
- Cai, Y.; Kong, F.; Shi, L.; Yu, Y. Spatial heterogeneity of cyanobacterial communities and genetic variation of Microcystis populations within large, shallow eutrophic lakes (Lake Tiahu and Lake Chaohu, China). J. Environ. Sci. 2012, 24, 1832–1842. [Google Scholar] [CrossRef]
- Otten, T.G.; Xu, H.; Qin, B.; Zhu, G.; Paerl, H.W. Spatiotemporal patterns and ecophysiology of toxigenic Microcystis blooms in Lake Taihu, China: Implications for water quality management. Environ. Sci. Technol. 2012, 46, 3480–3488. [Google Scholar] [CrossRef] [PubMed]
- Dyble, J.; Fahnenstiel, G.L.; Litaker, R.W.; Millie, D.F.; Tester, P.A. Microcystin concentrations and genetic diversity of Microcystis in the lower Great Lakes. Environ. Toxicol. 2008, 23, 507–516. [Google Scholar] [CrossRef]
- Lydolph, M.C.; Jacobsen, J.; Arctander, P.; Gilbert, M.T.P.; Gilichinsky, D.A.; Hansen, A.J.; Willerslev, E.; Lange, L. Beringian paleoecology inferred from permafrost-preserved fungal DNA. Appl. Environ. Microbiol. 2015, 71, 1012–1017. [Google Scholar] [CrossRef]
- Turner, C.R.; Barnes, M.A.; Xu, C.C.Y.; Jones, S.E.; Jerde, C.L.; Lodge, D.M. Particle size distribution and optimal capture of aqueous macrobial eDNA. Methods Ecol. Evol. 2014, 5, 676–684. [Google Scholar] [CrossRef]
- Al-Tebrineh, J.; Mihali, T.K.; Pomati, F.; Neilan, B.A. Detection of saxitoxin-producing cyanobacteria and Anabaena circinalis in environmental water blooms by quantitative PCR. Appl. Environ. Microbiol. 2010, 76, 7836–7842. [Google Scholar] [CrossRef]
- Vaitomaa, J.; Rantala, A.; Halinen, K.; Rouhiainen, L.; Tallberg, P.; Mokelke, L.; Sivonen, K. Quantitative real-time PCR for determination of microcystin synthetase E copy numbers of Microcystis and Anabaena in lakes. Appl. Environ. Microbiol. 2003, 69, 7289–7297. [Google Scholar] [CrossRef]
- Weber, A.A.-T.; Pawlowski, J. Can abundance of protists be inferred from sequence data: A case study of Foraminifera. PLoS ONE 2013, 8, e56739. [Google Scholar] [CrossRef]
- Weller, D.I. Detection, identification and toxigenicity of cyanobacteria in New Zealand lakes using PCR-based methods. N. Z. J. Mar. Freshw. Res. 2011, 45, 651–664. [Google Scholar] [CrossRef]
- Park, B.S.; Li, Z.; Kang, Y.-H.; Shin, H.H.; Joo, J.-H. Distinct bloom dynamics of toxic and non-toxic Microcystis (Cyanobacteria) subpopulations in Hoedong Reservoir (Korea). Microb. Ecol. 2018, 75, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Jia, R.; Liu, Y.; Zhang, H. Detection of microcystin-producing cyanobacteria in a reservoir by whole cell quantitative PCR. Procedia Environ. Sci. 2011, 10, 2272–2279. [Google Scholar] [CrossRef][Green Version]
- Feng, W.; Bulté, G.; Lougheed, S.C. Environmental DNA surveys help to identify winter hibernacula of a temperate freshwater turtle. Environ. DNA 2020, 2, 200–209. [Google Scholar] [CrossRef]
- Deiner, K.; Altermatt, F. Transport distance of invertebrate environmental DNA in a natural river. PLoS ONE 2014, 9, e88786. [Google Scholar] [CrossRef]
- Canadian Phycological Culture Centre List of Cultures. Available online: https://uwaterloo.ca/canadian-phycological-culture-centre/sites/ca.canadian-phycological-culture-centre/files/uploads/files/cpcc_list_of_cultures_nov_20_13.pdf (accessed on 1 June 2019).
- Ichimura, T. Isolation and culture methods of algae. In Methods in Phycological Studies; Nishizawa, K., Chihara, M., Eds.; Kyoritsu Shuppan: Tokyo, Japan, 1979; p. 301. (In Japanese) [Google Scholar]
- Neilan, B.A.; Jacobs, D.; Del Dot, T.; Blackall, L.L.; Hawkins, P.R.; Cox, P.T.; Goodman, A.E. rRNA sequences and evolutionary relationships among toxic and nontoxic cyanobacteria of the genus Microcystis. Int. J. Syst. Evol. Microbiol. 1997, 47, 693–697. [Google Scholar] [CrossRef]
- Sipari, H.; Rantala-Ylinen, A.; Jokela, J.; Oksanen, I.; Sivonen, K. Development of a chip assay and quantitative PCR for detecting microcystin synthetase E gene expression. Appl. Environ. Microbiol. 2010, 76, 3797–3805. [Google Scholar] [CrossRef]
- Kitchens, C.M.; Johengen, T.H.; Davis, T.W. Establishing spatial and temporal patterns in Microcystis sediment seed stock viability and their relationship to subsequent bloom development in western Lake Erie. PLoS ONE 2018, 13, e0206821. [Google Scholar] [CrossRef]
- Ma, J.; Qin, B.; Paerl, H.W.; Brookes, J.D.; Hall, N.S.; Shi, K.; Zhou, Y.; Guo, J.; Li, Z.; Xu, H.; et al. The persistence of cyanobacterial (Microcystis spp.) blooms throughout winter in Lake Taihu, China. Limnol. Oceanogr. 2016, 61, 711–722. [Google Scholar] [CrossRef]
- Misson, B.; Sabart, M.; Amblard, C.; Latour, D. Benthic survival of Microcystis: Long-term viability and ability to transcribe microcystin genes. Harmful Algae 2012, 13, 20–25. [Google Scholar] [CrossRef]
- Cires, S.; Wormer, L.; Agha, R.; Quesada, A. Overwintering populations of Anabaena, Aphanizomenon and Microcystis as potential inocula for summer blooms. J. Plankton Res. 2013, 35, 1254–1266. [Google Scholar] [CrossRef]
- Bach, H.-J.; Tomanova, J.; Schloter, M.; Munch, J.C. Enumeration of total bacteria and bacteria with genes for proteolytic activity in pure cultures and in environmental samples by quantitative PCR mediate amplification. J. Microbiol. Methods 2002, 49, 235–245. [Google Scholar] [CrossRef]
- Ouellette, A.J.A.; Wilhelm, S.W. Toxic cyanobacteria: The evolving molecular toolbox. Front. Ecol. Environ. 2003, 1, 359–366. [Google Scholar] [CrossRef]
- Jin, J. Detecting the Invisible: Early Monitoring of Cyanobacterial Harmful Algal Blooms with Quantitative PCR. Master’s Thesis, Queen’s University, Kingston, ON, Canada, April 2020. [Google Scholar]
- Yuan, J.; Kim, H.-J.; Filstrup, C.T.; Guo, B.; Imerman, P.; Ensley, S.; Yoon, K.-J. Utility of a PCR-based method for rapid and specific detection of toxigenic Microcystis spp. in farm ponds. J. Vet. Diagn. Investig. 2020, 32, 369–381. [Google Scholar] [CrossRef]
- Chiu, Y.-T.; Chen, Y.-H.; Wang, T.-S.; Yen, H.-K.; Lin, T.-F. A qPCR-based tool to diagnose the presence of harmful cyanobacteria and cyanotoxins in drinking water sources. Int. J. Environ. Res. Public Health 2017, 14, 547. [Google Scholar] [CrossRef]
- Cao, H.-S.; Tao, Y.; Kong, F.-X.; Yang, Z. Relationship between temperature and cyanobacterial recruitment from sediments in laboratory and field studies. J. Freshw. Ecol. 2008, 23, 405–412. [Google Scholar] [CrossRef]
- Liu, X.; Lu, X.; Chen, Y. The effects of temperature and nutrient ratios on Microcystis blooms in Lake Taihu, China: An 11-year investigation. Harmful Algae 2011, 10, 337–343. [Google Scholar] [CrossRef]
- Thomas, R.H.; Walsby, A.E. The effect of temperature on recovery of buoyancy by Microcystis. J. Gen. Microbiol. 1986, 132, 1665–1672. [Google Scholar] [CrossRef]
- Stahl-Delbanco, A.; Hansson, L.-A. Effects of bioturbation on recruitment of algal cells from the “seed bank” of lake sediments. Limnol. Oceanogr. 2002, 47, 1836–1843. [Google Scholar] [CrossRef]
- Scott, J.T.; Myer, G.E.; Stewart, R.; Walther, E.G. On the mechanism of Langmuir circulations and their role in epilimnion mixing. Limnol. Oceanogr. 1969, 14, 493–503. [Google Scholar] [CrossRef]
- Blottiere, L. The Effects of Wind-Induced Mixing on the Structure and Functioning of Shallow Freshwater Lakes in a Context of Global Change. Ph.D. Thesis, Université Paris-Saclay, Paris, France, 2015. [Google Scholar]
- Deming, J.W.; Young, J.N. The role of exopolysaccharides in microbial adaptation to cold habitats. In Psychrophiles: From Biodiversity to Biotechnology, 2nd ed.; Margesin, R., Schinner, F., Marx, J.-C., Gerday, C., Eds.; Springer: Berlin, Germany, 2017; pp. 259–284. [Google Scholar]
- Watanabe, M.F.; Oishi, S. Effects of environmental factors on toxicity of a cyanobacterium (Microcystis aeruginosa) under culture conditions. Appl. Environ. Microbiol. 1985, 49, 1342–1344. [Google Scholar] [CrossRef] [PubMed]
- Walls, J.T.; Wyatt, K.H.; Doll, J.C.; Rubenstein, E.M.; Rober, A.R. Hot and toxic: Temperature regulates microcystin release from cyanobacteria. Sci. Total Environ. 2018, 1, 610–611. [Google Scholar] [CrossRef] [PubMed]
- Dziallas, C.; Grossart, H.-P. Increasing oxygen radicals and water temperature select for toxic Microcystis sp. PLoS ONE 2011, 6, e25569. [Google Scholar] [CrossRef]
- Zilliges, Y.; Kehr, J.; Meissner, S.; Ishida, K.; Mikkat, S.; Hagemann, M.; Kaplan, A.; Borner, T.; Dittman, E. The cyanobacteria hepatotoxin microcystin binds to proteins and increases the fitness of Microcystis under oxidative stress conditions. PLoS ONE 2011, 6, e17615. [Google Scholar] [CrossRef] [PubMed]
- Perakis, S.S.; Welch, E.B.; Jacoby, J.M. Sediment-to-water blue-green algal recruitment in response to alum and environmental factors. Hydrobiologia 1996, 318, 165–177. [Google Scholar] [CrossRef]
- Zhang, M.; Yang, Z.; Yu, Y.; Shi, X. Interannual and seasonal shift between Microcystis and Dolichospermum: A 7-year investigation in Lake Chaohu, China. Water 2020, 12, 1978. [Google Scholar] [CrossRef]





| Target | Primer | Sequence (5′ to 3′) | Size | Reference |
|---|---|---|---|---|
| Microcystis 16S gene | MIC 16S-F | GCCGCRAGGTGAAAMCTAA | 220 bp | [50] |
| MIC 16S-R | AATCCAAARACCTTCCTCCC | |||
| Microcystis mcyE gene | MIC mcyE-F | AAGCAAACTGCTCCCGGTATC | 120 bp | [51] |
| MIC mcyE-R | CAATGGGAGCATAACGAGTCAA |
| Factor | Df | F | P | Wilks’ Partial ηp2 |
|---|---|---|---|---|
| (A) Concentrations of M. aeruginosa 16S rRNA gene | ||||
| Sampling time | 5 | 6.25 | <0.001 | 0.201 |
| Water column level (near-surface, near-bottom, bottom) | 1 | 308.12 | <0.001 | 0.713 |
| Depth | 1 | 9.70 | =0.002 | 0.073 |
| Sampling time × depth | 5 | 5.66 | <0.001 | 0.186 |
| Water column level × depth | 1 | 81.63 | <0.001 | 0.397 |
| Error | 124 | |||
| (B) Concentrations of M. aeruginosa mcyE gene | ||||
| Sampling time | 5 | 7.48 | <0.001 | 0.232 |
| Water column level (near-surface, near-bottom) | 1 | 323.39 | <0.001 | 0.723 |
| Depth | 1 | 10.67 | =0.001 | 0.079 |
| Sampling time × depth | 5 | 6.15 | <0.001 | 0.199 |
| Water column level × depth | 1 | 86.54 | <0.001 | 0.411 |
| Error | 124 | |||
| Gene | Coefficient | Value | Std. Error |
|---|---|---|---|
| 16S rRNA | A1 | 0.02924 | 0.00382 |
| A2 | 0.80606 | 0.07893 | |
| X0 | 14.7098 | 0.49923 | |
| p | 7.19072 | 0.69382 | |
| mcyE | A1 | 0.03037 | 0.00365 |
| A2 | 0.77372 | 0.06673 | |
| X0 | 14.4735 | 0.43581 | |
| p | 7.41774 | 0.68911 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, H.; Jin, J.; Chen, B.; Lefebvre, D.D.; Lougheed, S.C.; Wang, Y. Depth-Dependent Spatiotemporal Dynamics of Overwintering Pelagic Microcystis in a Temperate Water Body. Microorganisms 2021, 9, 1718. https://doi.org/10.3390/microorganisms9081718
Tian H, Jin J, Chen B, Lefebvre DD, Lougheed SC, Wang Y. Depth-Dependent Spatiotemporal Dynamics of Overwintering Pelagic Microcystis in a Temperate Water Body. Microorganisms. 2021; 9(8):1718. https://doi.org/10.3390/microorganisms9081718
Chicago/Turabian StyleTian, Haolun, Junjie Jin, Bojian Chen, Daniel D. Lefebvre, Stephen C. Lougheed, and Yuxiang Wang. 2021. "Depth-Dependent Spatiotemporal Dynamics of Overwintering Pelagic Microcystis in a Temperate Water Body" Microorganisms 9, no. 8: 1718. https://doi.org/10.3390/microorganisms9081718
APA StyleTian, H., Jin, J., Chen, B., Lefebvre, D. D., Lougheed, S. C., & Wang, Y. (2021). Depth-Dependent Spatiotemporal Dynamics of Overwintering Pelagic Microcystis in a Temperate Water Body. Microorganisms, 9(8), 1718. https://doi.org/10.3390/microorganisms9081718





