The Ever-Expanding Pseudomonas Genus: Description of 43 New Species and Partition of the Pseudomonas putida Group
Abstract
:1. Introduction
2. Materials and Methods
2.1. Pseudomonas Strains
2.2. Genome Sequencing, Assembly, and Functional Annotation
2.3. Taxonomic Affiliations and Phylogenetic Analyses
2.4. Cyclic Lipopeptide (CLP) NRPS Analysis
3. Results and Discussion
3.1. Defining New Pseudomonas Species
3.2. Comparison of Whole Genome and rpoD-based Phylogenies
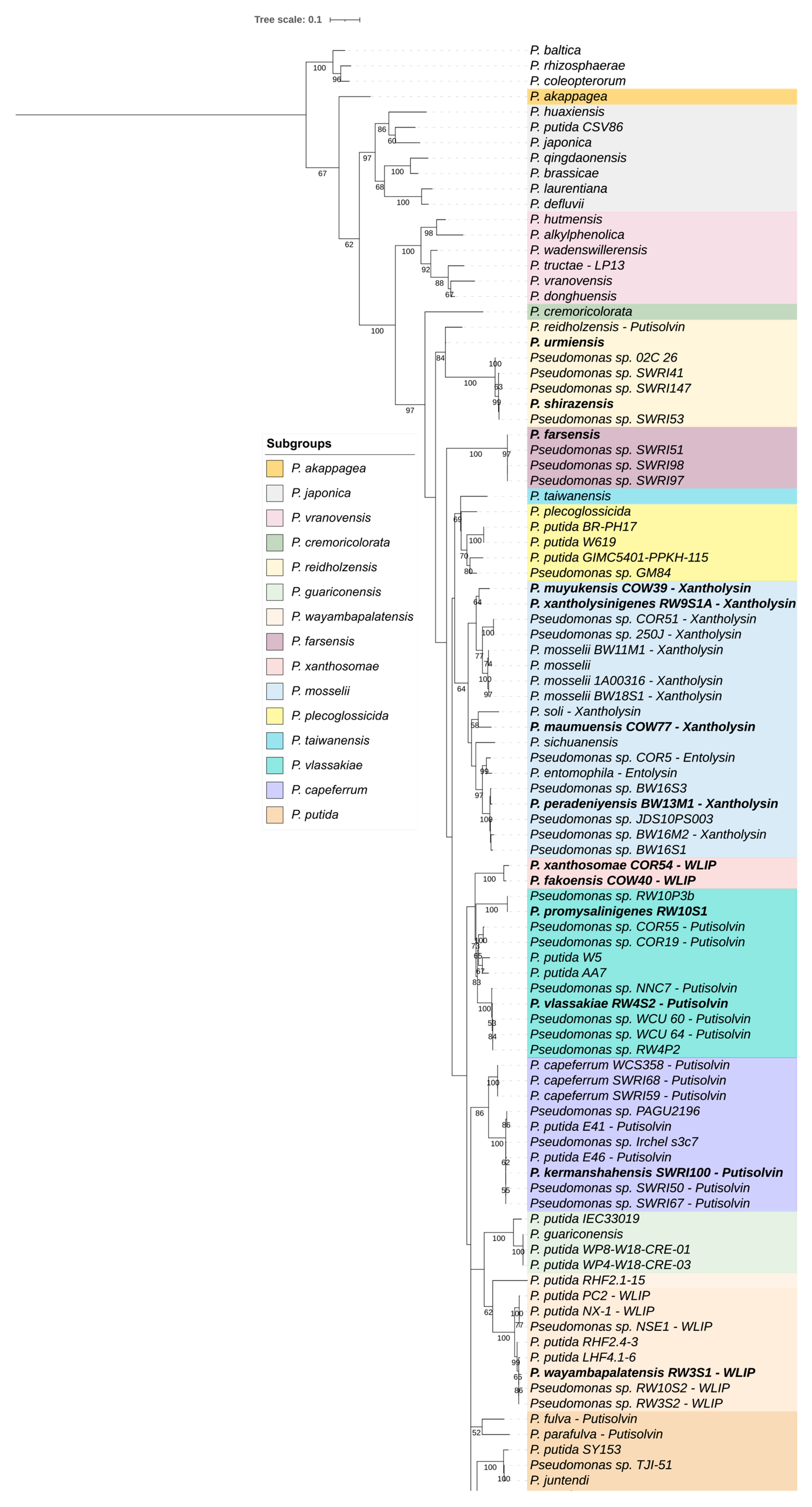
3.3. Genomic Diversity within the P. putida Group
3.3.1. Identification and Reassignment at the Species Level
3.3.2. Distribution of CLP biosynthesis Gene Clusters
3.3.3. Partitioning of the P. putida Group
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A. Descriptions of the 43 New Pseudomonas Species
References
- Stackebrandt, E. Report of the Ad Hoc Committee for the Re-Evaluation of the Species Definition in Bacteriology. Int. J. Syst. Evol. Microbiol. 2002, 52, 1043–1047. [Google Scholar] [CrossRef] [PubMed]
- Pallen, M.J.; Telatin, A.; Oren, A. The Next Million Names for Archaea and Bacteria. Trends Microbiol. 2021, 29, 289–298. [Google Scholar] [CrossRef]
- Gevers, D.; Cohan, F.M.; Lawrence, J.G.; Spratt, B.G.; Coenye, T.; Feil, E.J.; Stackebrandt, E.; de Peer, Y.V.; Vandamme, P.; Thompson, F.L.; et al. Re-Evaluating Prokaryotic Species. Nat. Rev. Microbiol. 2005, 3, 733–739. [Google Scholar] [CrossRef]
- Gomila, M.; Peña, A.; Mulet, M.; Lalucat, J.; García-Valdés, E. Phylogenomics and Systematics in Pseudomonas. Front. Microbiol. 2015, 6, 214. [Google Scholar] [CrossRef] [Green Version]
- Mulet, M.; Lalucat, J.; García-Valdés, E. DNA Sequence-Based Analysis of the Pseudomonas Species. Environ. Microbiol. 2010, 12, 1513–1530. [Google Scholar] [CrossRef] [Green Version]
- Lalucat, J.; Mulet, M.; Gomila, M.; García-Valdés, E. Genomics in Bacterial Taxonomy: Impact on the Genus Pseudomonas. Genes 2020, 11, 139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawabe, T.; Ogura, Y.; Matsumura, Y.; Feng, G.; Amin, A.R.; Mino, S.; Nakagawa, S.; Sawabe, T.; Kumar, R.; Fukui, Y.; et al. Updating the Vibrio Clades Defined by Multilocus Sequence Phylogeny: Proposal of Eight New Clades, and the Description of Vibrio Tritonius sp. Nov. Front. Microbiol. 2013, 4, 414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.; Oh, H.-S.; Park, S.-C.; Chun, J. Towards a Taxonomic Coherence between Average Nucleotide Identity and 16S RRNA Gene Sequence Similarity for Species Demarcation of Prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome Sequence-Based Species Delimitation with Confidence Intervals and Improved Distance Functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meier-Kolthoff, J.P.; Klenk, H.-P.; Göker, M. Taxonomic Use of DNA G+C Content and DNA–DNA Hybridization in the Genomic Age. Int. J. Syst. Evol. Microbiol. 2014, 64, 352–356. [Google Scholar] [CrossRef] [PubMed]
- Auch, A.F.; von Jan, M.; Klenk, H.-P.; Göker, M. Digital DNA-DNA Hybridization for Microbial Species Delineation by Means of Genome-to-Genome Sequence Comparison. Stand. Genom. Sci. 2010, 2, 117–134. [Google Scholar] [CrossRef] [Green Version]
- Chaumeil, P.-A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A Toolkit to Classify Genomes with the Genome Taxonomy Database. Bioinformatics 2019, 36, btz848. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Chuvochina, M.; Waite, D.W.; Rinke, C.; Skarshewski, A.; Chaumeil, P.-A.; Hugenholtz, P. A Standardized Bacterial Taxonomy Based on Genome Phylogeny Substantially Revises the Tree of Life. Nat. Biotechnol. 2018, 36, 996–1004. [Google Scholar] [CrossRef]
- Moore, E.R.B.; Mau, M.; Arnscheidt, A.; Böttger, E.C.; Hutson, R.A.; Collins, M.D.; Van De Peer, Y.; De Wachter, R.; Timmis, K.N. The Determination and Comparison of the 16S RRNA Gene Sequences of Species of the Genus Pseudomonas (Sensu Stricto and Estimation of the Natural Intrageneric Relationships. Syst. Appl. Microbiol. 1996, 19, 478–492. [Google Scholar] [CrossRef]
- Brosch, R.; Lefèvre, M.; Grimont, F.; Grimont, P.A.D. Taxonomic Diversity of Pseudomonads Revealed by Computer-Interpretation of Ribotyping Data. Syst. Appl. Microbiol. 1996, 19, 541–555. [Google Scholar] [CrossRef]
- Mulet, M.; García-Valdés, E.; Lalucat, J. Phylogenetic Affiliation of Pseudomonas putida Biovar A and B Strains. Res. Microbiol. 2013, 164, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Girard, L.; Lood, C.; Rokni-Zadeh, H.; van Noort, V.; Lavigne, R.; De Mot, R. Reliable Identification of Environmental Pseudomonas Isolates Using the RpoD Gene. Microorganisms 2020, 8, 1166. [Google Scholar] [CrossRef]
- Hesse, C.; Schulz, F.; Bull, C.T.; Shaffer, B.T.; Yan, Q.; Shapiro, N.; Hassan, K.A.; Varghese, N.; Elbourne, L.D.H.; Paulsen, I.T.; et al. Genome-Based Evolutionary History of Pseudomonas Spp. Environ. Microbiol. 2018, 20, 2142–2159. [Google Scholar] [CrossRef] [PubMed]
- Höfte, M.; De Vos, P. Plant pathogenic Pseudomonas species. In Plant-Associated Bacteria; Gnanamanickam, S.S., Ed.; Springer: Dordrecht, The Netherlands, 2007; pp. 507–533. ISBN 978-1-4020-4536-3. [Google Scholar]
- Wiklund, T. Pseudomonas Anguilliseptica Infection as a Threat to Wild and Farmed Fish in the Baltic Sea. Microbiol. Aust. 2016, 37, 135. [Google Scholar] [CrossRef] [Green Version]
- Beaton, A.; Lood, C.; Cunningham-Oakes, E.; MacFadyen, A.; Mullins, A.J.; Bestawy, W.E.; Botelho, J.; Chevalier, S.; Coleman, S.; Dalzell, C.; et al. Community-Led Comparative Genomic and Phenotypic Analysis of the Aquaculture Pathogen Pseudomonas Baetica A390T Sequenced by Ion Semiconductor and Nanopore Technologies. FEMS Microbiol. Lett. 2018, 365. [Google Scholar] [CrossRef] [Green Version]
- Wasi, S.; Tabrez, S.; Ahmad, M. Use of Pseudomonas spp. for the Bioremediation of Environmental Pollutants: A Review. Environ. Monit. Assess. 2013, 185, 8147–8155. [Google Scholar] [CrossRef]
- Weller, D.M. Pseudomonas Biocontrol Agents of Soilborne Pathogens: Looking Back Over 30 Years. Phytopathology 2007, 97, 250–256. [Google Scholar] [CrossRef] [Green Version]
- Viggor, S.; Jõesaar, M.; Vedler, E.; Kiiker, R.; Pärnpuu, L.; Heinaru, A. Occurrence of Diverse Alkane Hydroxylase AlkB Genes in Indigenous Oil-Degrading Bacteria of Baltic Sea Surface Water. Mar. Pollut. Bull. 2015, 101, 507–516. [Google Scholar] [CrossRef]
- Gwon, H.-J.; Teruhiko, I.; Shigeaki, H.; Baik, S.-H. Identification of Novel Non-Metal Haloperoxidases from the Marine Metagenome. J. Microbiol. Biotechnol. 2014, 24, 835–842. [Google Scholar] [CrossRef] [PubMed]
- Mendes, R.; Kruijt, M.; de Bruijn, I.; Dekkers, E.; van der Voort, M.; Schneider, J.H.M.; Piceno, Y.M.; DeSantis, T.Z.; Andersen, G.L.; Bakker, P.A.H.M.; et al. Deciphering the Rhizosphere Microbiome for Disease-Suppressive Bacteria. Science 2011, 332, 1097–1100. [Google Scholar] [CrossRef] [PubMed]
- Gross, H.; Loper, J.E. Genomics of Secondary Metabolite Production by Pseudomonas spp. Nat. Prod. Rep. 2009, 26, 1408–1446. [Google Scholar] [CrossRef]
- Götze, S.; Stallforth, P. Structure Elucidation of Bacterial Nonribosomal Lipopeptides. Org. Biomol. Chem. 2020, 18, 1710–1727. [Google Scholar] [CrossRef] [PubMed]
- Geudens, N.; Martins, J.C. Cyclic Lipodepsipeptides From Pseudomonas spp.–Biological Swiss-Army Knives. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Oni, F.E.; Geudens, N.; Omoboye, O.O.; Bertier, L.; Hua, H.G.K.; Adiobo, A.; Sinnaeve, D.; Martins, J.C.; Höfte, M. Fluorescent Pseudomonas and Cyclic Lipopeptide Diversity in the Rhizosphere of Cocoyam (Xanthosoma sagittifolium). Environ. Microbiol. 2019, 21, 1019–1034. [Google Scholar] [CrossRef] [Green Version]
- Oni, F.E.; Geudens, N.; Onyeka, J.T.; Olorunleke, O.F.; Salami, A.E.; Omoboye, O.O.; Arias, A.A.; Adiobo, A.; De Neve, S.; Ongena, M.; et al. Cyclic Lipopeptide—Producing Pseudomonas Koreensis Group Strains Dominate the Cocoyam Rhizosphere of a Pythium Root Rot Suppressive Soil Contrasting with P. putida Prominence in Conducive Soils. Environ. Microbiol. 2020, 22, 5137–5155. [Google Scholar] [CrossRef]
- Lood, C.; Peeters, C.; Lamy-Besnier, Q.; Wagemans, J.; De Vos, D.; Proesmans, M.; Pirnay, J.-P.; Echahidi, F.; Piérard, D.; Thimmesch, M.; et al. Genomics of an Endemic Cystic Fibrosis Burkholderia Multivorans Strain Reveals Low Within-Patient Evolution but High between-Patient Diversity. PLoS Pathog. 2021, 17, e1009418. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- De Coster, W.; D’Hert, S.; Schultz, D.T.; Cruts, M.; Van Broeckhoven, C. NanoPack: Visualizing and Processing Long-Read Sequencing Data. Bioinforma. Oxf. Engl. 2018, 34, 2666–2669. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving Bacterial Genome Assemblies from Short and Long Sequencing Reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality Assessment Tool for Genome Assemblies. Bioinforma. Oxf. Engl. 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI Prokaryotic Genome Annotation Pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Pritchard, L.; Glover, R.H.; Humphris, S.; Elphinstone, J.G.; Toth, I.K. Genomics and Taxonomy in Diagnostics for Food Security: Soft-Rotting Enterobacterial Plant Pathogens. Anal. Methods 2016, 8, 12–24. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [Green Version]
- Wittouck, S.; Wuyts, S.; Meehan, C.J.; Van Noort, V.; Lebeer, S. A Genome-Based Species Taxonomy of the Lactobacillus Genus Complex. mSystems 2019, 4, e00264-19. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. AntiSMASH 6.0: Improving Cluster Detection and Comparison Capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Götze, S.; Stallforth, P. Structure, Properties, and Biological Functions of Nonribosomal Lipopeptides from Pseudomonads. Nat. Prod. Rep. 2019, 37, 29–54. [Google Scholar] [CrossRef]
- Girard, L.; Höfte, M.; Mot, R.D. Lipopeptide Families at the Interface between Pathogenic and Beneficial Pseudomonas-Plant Interactions. Crit. Rev. Microbiol. 2020, 1–23. [Google Scholar] [CrossRef]
- Morimoto, Y.; Tohya, M.; Aibibula, Z.; Baba, T.; Daida, H.; Kirikae, T. Re-Identification of Strains Deposited as Pseudomonas aeruginosa, Pseudomonas fluorescens and Pseudomonas putida in GenBank Based on Whole Genome Sequences. Int. J. Syst. Evol. Microbiol. 2020, 70, 5958–5963. [Google Scholar] [CrossRef]
- Tohya, M.; Watanabe, S.; Tada, T.; Tin, H.H.; Kirikae, T. Genome Analysis-Based Reclassification of Pseudomonas fuscovaginae and Pseudomonas shirazica as Later Heterotypic Synonyms of Pseudomonas asplenii and Pseudomonas asiatica, Respectively. Int. J. Syst. Evol. Microbiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Omoboye, O.O.; Oni, F.E.; Batool, H.; Yimer, H.Z.; De Mot, R.; Höfte, M. Pseudomonas Cyclic Lipopeptides Suppress the Rice Blast Fungus Magnaporthe Oryzae by Induced Resistance and Direct Antagonism. Front. Plant Sci. 2019, 10, 901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, D.D.; Melnik, A.V.; Koyama, N.; Lu, X.; Schorn, M.; Fang, J.; Aguinaldo, K.; Lincecum, T.L.; Ghequire, M.G.K.; Carrion, V.J.; et al. Indexing the Pseudomonas Specialized Metabolome Enabled the Discovery of Poaeamide B and the Bananamides. Nat. Microbiol. 2016, 2, 16197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vallet-Gely, I.; Novikov, A.; Augusto, L.; Liehl, P.; Bolbach, G.; Péchy-Tarr, M.; Cosson, P.; Keel, C.; Caroff, M.; Lemaitre, B. Association of Hemolytic Activity of Pseudomonas entomophila, a Versatile Soil Bacterium, with Cyclic Lipopeptide Production. Appl. Environ. Microbiol. 2010, 76, 910–921. [Google Scholar] [CrossRef] [Green Version]
- Pascual, J.; García-López, M.; Carmona, C.; Sousa, T.d.S.; de Pedro, N.; Cautain, B.; Martín, J.; Vicente, F.; Reyes, F.; Bills, G.F.; et al. Pseudomonas soli sp. Nov., a Novel Producer of Xantholysin Congeners. Syst. Appl. Microbiol. 2014, 37, 412–416. [Google Scholar] [CrossRef]
- Aiman, S.; Shehroz, M.; Munir, M.; Gul, S.; Shah, M.; Khan, A. Species-Wide Genome Mining of Pseudomonas putida for Potential Secondary Metabolites and Drug-Like Natural Products Characterization. J. Proteomics Bioinform. 2018, 11. [Google Scholar] [CrossRef]
- Rokni-Zadeh, H.; Li, W.; Sanchez-Rodriguez, A.; Sinnaeve, D.; Rozenski, J.; Martins, J.C.; De Mot, R. Genetic and Functional Characterization of Cyclic Lipopeptide White-Line-Inducing Principle (WLIP) Production by Rice Rhizosphere Isolate Pseudomonas Putida RW10S2. Appl. Environ. Microbiol. 2012, 78, 4826–4834. [Google Scholar] [CrossRef] [Green Version]
- Bernat, P.; Nesme, J.; Paraszkiewicz, K.; Schloter, M.; Płaza, G. Characterization of Extracellular Biosurfactants Expressed by a Pseudomonas putida Strain Isolated from the Interior of Healthy Roots from Sida Hermaphrodita Grown in a Heavy Metal Contaminated Soil. Curr. Microbiol. 2019, 76, 1320–1329. [Google Scholar] [CrossRef]
- Kuiper, I.; Lagendijk, E.L.; Pickford, R.; Derrick, J.P.; Lamers, G.E.M.; Thomas-Oates, J.E.; Lugtenberg, B.J.J.; Bloemberg, G.V. Characterization of Two Pseudomonas putida Lipopeptide Biosurfactants, Putisolvin I and II, Which Inhibit Biofilm Formation and Break down Existing Biofilms. Mol. Microbiol. 2004, 51, 97–113. [Google Scholar] [CrossRef]
- Dubern, J.-F.; Coppoolse, E.R.; Stiekema, W.J.; Bloemberg, G.V. Genetic and Functional Characterization of the Gene Cluster Directing the Biosynthesis of Putisolvin I and II in Pseudomonas putida Strain PCL1445. Microbiol. Read. Engl. 2008, 154, 2070–2083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Rokni-Zadeh, H.; De Vleeschouwer, M.; Ghequire, M.G.K.; Sinnaeve, D.; Xie, G.-L.; Rozenski, J.; Madder, A.; Martins, J.C.; De Mot, R. The Antimicrobial Compound Xantholysin Defines a New Group of Pseudomonas Cyclic Lipopeptides. PLoS ONE 2013, 8, e62946. [Google Scholar] [CrossRef] [Green Version]
- Molina-Santiago, C.; Udaondo, Z.; Ramos, J.-L. Draft Whole-Genome Sequence of the Antibiotic-Producing Soil Isolate Pseudomonas sp. Strain 250J. Environ. Microbiol. Rep. 2015, 7, 288–292. [Google Scholar] [CrossRef] [PubMed]
- Biessy, A.; Novinscak, A.; Blom, J.; Léger, G.; Thomashow, L.S.; Cazorla, F.M.; Josic, D.; Filion, M. Diversity of Phytobeneficial Traits Revealed by Whole-Genome Analysis of Worldwide-Isolated Phenazine-Producing Pseudomonas Spp. Environ. Microbiol. 2019, 21, 437–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Groups/Subgroups | Species/Subspecies | Total No. of Species/ Subspecies | rpoD Identity (%) 1 | ANIb (%) 1 |
|---|---|---|---|---|
| Existing Groups | ||||
| P. aeruginosa G | P. aeruginosa, P. citronellolis, P. delhiensis, P. humi, P. jinjuensis, P. knackmussii, P. nicosulfuronedens, P. nitritireducens, P. nitroreducens, P. panipatensis | 10 | 77.85–95.98 | 80.57–94.48 |
| P. anguilliseptica G | P. anguilliseptica, P. benzenivorans, P. borbori, P. cuatrocienegasensis, P. guineae, P. leptonychotis, P. marincola, P. peli, P. segetis, P. taeanensis | 10 | 72.33–91.58 | 76.68–89.45 |
| P. fluorescens G | 134 | – | – | |
| P. asplenii SG | P. agarici, P. asplenii, P. batumici, P. fuscovaginae, P. vanderleydeniana | 5 | 84.41–89.66 | 84.10–88.35 |
| P. chlororaphis SG | P. chlororaphis subsp. aurantiaca, P. chlororaphis subsp. aureofaciens, P. chlororaphis subsp. chlororaphis, P. chlororaphis subsp. piscium | 4 | 97.83–98.45 | 94.73–96.95 |
| P. corrugata SG | P. alvandae, P. beijieensis, P. brassicacearum, P. corrugata, P. kilonensis, P. marvdashtae, P. mediterranea, P. tehranensis, P. thivervalensis, P. viciae, P. zanjanensis, P. zarinae | 12 | 89.75–97.36 | 85.55–95.75 |
| P. fluorescens SG | P. allii, P. antartica, P. asgharzadehiana, P. aylmerense, P. azadiae, P. azotoformans, P. canadensis, P. carnis, P. cedrina subsp. cedrina, P. cedrina subsp. fulgida, P. costantinii, P. cremoris, P. cyclaminis, P. edaphica, P. extremaustralis, P. extremorientalis, P. fildesensis, P. fluorescens, P. grimontii, P. haemolytica, P. kairouanesis, P. karstica, P. khavaziana, P. kitaguniensis, P. lactis, P. libanensis, P. lurida, P. marginalis, P. nabeulensis, P. orientalis, P. palleroniana, P. panacis, P. paracarnis, P. paralactis, P. pisciculturae, P. poae, P. rhodesiae, P. salmasensis, P. salomonii, P. simiae, P. sivasensis, P. spelaei, P. synxantha, P. tolaasii, P. tritici, P. trivialis, P. veronii, P. yamanorum | 48 | 85.16–98.91 | 83.52–95.68 |
| P. fragi SG | P. bubulae, P. deceptionensis, P. endophytica, P. fragi, P. helleri, P. lundensis, P. psychrophila, P. saxonica, P. taetrolens, P. versuta, P. weihenstephanensis | 11 | 83.00–97.67 | 80.50–90.49 |
| P. gessardii SG | P. brennerii, P. gessardii, P. mucidolens, P. proteolytica, P. shahriarae | 5 | 90.57–97.53 | 85.57–92.54 |
| P. jessenii SG | P. asgharzadehiana, P. azerbaijanoccidens, P. izuensis, P. jessenii, P. laurylsulfatiphila, P. laurylsulfatovorans, P. mohnii, P. moorei, P. reinekei, P. umsongensis, P. vancouverensis | 11 | 90.37–100 | 84.91–95.51 |
| P. koreensis SG | P. atacamensis, P. atagosis, P. baetica, P. bananamidigenes, P. botevensis, P. crudilactis, P. ekonensis, P. glycinae, P. granadensis, P. hamedanensis, P. helmanticensis, P. iranensis, P. khorasanensis, P. koreensis, P. kribbensis, P. monsensis, P. moraviensis, P. neuropathica, P. siliginis, P. tensinigenes, P. triticicola, P. zeae | 22 | 85.40–99.53 | 82.48–96.09 |
| P. mandelii SG | P. arsenicoxydans, P. farris, P. frederiksbergensis, P. gregormendelii, P. lini, P. mandelii, P. migulae, P. mucoides, P. piscium, P. prosekii, P. silesiensis | 11 | 91.04–96.89 | 84.68–94.29 |
| P. protegens SG | P. aestus, P. protegens, P. saponiphila, P. sessilinigenes | 4 | 89.52–95.57 | 86.41–91.86 |
| P. kielensis SG | P. kielensis | 1 | – | – |
| P. linyingensis G | P. guangdongensis, P. linyingensis, P. oryzae, P. sagittaria | 4 | 79.94–93.85 | 85.19–92.01 |
| P. lutea G | P. abietaniphila, P. bohemica, P. graminis, P. lutea | 4 | 83.31–88.30 | 81.89–85.81 |
| P. oleovorans G | P. alcaliphila, P. chaetoceroseae, P. chengduensis, P. composti, P. guguanensis, P. hydrolytica, P. indoloxydans, P. khazarica, P. mendocina, P. oleovorans, P. pseudoalcaligenes, P. sediminis, P. sihuisensis, P. toyotomiensis | 14 | 88.51–98.76 | 86.06–95.79 |
| P. oryzihabitans G | P. asuensis, P. duriflava, P. luteola, P. oryzihabitans, P. psychrotolerans, P. rhizoryzae, P. zeshuii | 7 | 66.46–94.18 | 73.64–88.62 |
| P. pertucinogena G | P. abyssi, P. aestusnigri, P. bauzanensis, P. formosensis, P. gallaeciencis, P. jilinensis, P. litoralis, P. oceani, P. pachastrellae, P. pelagia, P. pertucinogena, P. phragmitis, P. populi, P. profundi, P. sabulinigri, P. salegens, P. salina, P. saliphila, P. saudimassiliensis, P. xiamenensis, P. xinjiangensis, P. yangmingensis | 22 | 64.53–92.98 | 74.65–89.65 |
| P. putida G | 51 | |||
| P. akappagea SG | P. akappagea | 1 | – | – |
| P. japonica SG | P. brassicae, P. defluvii, P. huaxiensis, P. japonica, P. laurentiana, P. qingdaonensis | 6 | 82.69–95.05 | 80.96–91.58 |
| P. vranovensis SG | P. alkylphenolica, P. donghuensis, P. hutmensis, P. tructae, P. vranovensis, P. wadenswillerensis | 6 | 84.23–94.10 | 84.52–93.03 |
| P. cremoricolorata SG | P. cremoricolorata | 1 | – | – |
| P. reidholzensis SG | P. reidholzensis, P. shirazensis, P. urmiensis | 3 | 85.78–92.89 | 84.27–86.77 |
| P. guariconensis SG | P. guariconensis | 1 | – | – |
| P. wayambapalatensis SG | P. wayambapalatensis | 1 | – | – |
| P. farsensis SG | P. farsensis | 1 | – | – |
| P. xanthosomae SG | P. fakonensis, P. xanthosomae | 2 | 97.84 | 95.06 |
| P. mosselii SG | P. entomophila, P. maumuensis, P. mosselii, P. muyukensis, P. peradeniyensis, P. sichuanensis, P. soli, P. xantholysinigenes | 8 | 87.48–95.35 | 87.35–94.87 |
| P. taiwanensis SG | P. taiwanensis | 1 | – | – |
| P. plecoglossicida SG | P. plecoglossicida | 1 | – | – |
| P. vlassakiae SG | P. hunanensis, P. promysalinigenes, P. vlassakiae | 3 | 89.34–93.04 | 86.58 |
| P. capeferrum SG | P. capeferrum, P. kermanshahensis | 2 | 93.01 | 90.26 |
| P. putida SG | P. alloputida, P. anuradhapurensis, P. asiatica, P. fulva, P. inefficax, P. juntendi, P. kurunegalensis, P. monteilii, P. oryzicola, P. parafulva, P. putida, P. pyomelaminifaciens, P. persica, P. shirazica | 14 | 85.32–97.99 | 82.66–95.79 |
| P. resinovorans G | P. furukawaii, P. lalkuanensis, P. mangiferae, P. otitidis, P. resinovorans | 5 | 81.47–90.46 | 79.25–87.84 |
| P. straminea G | P. argentinensis, P. daroniae, P. dryadis, P. flavescens, P. punonensis, P. seleniipraecipitans, P. straminea | 7 | 86.02–93.63 | 82.83–88.54 |
| P. stutzeri G | P. azotofigens, P. balearica, P. chloritidismutans, P. kirkiae, P. kunmingensis, P. nitritititolerans, P. nosocomialis, P. perfectomarina, P. saudiphocaensis, P. songnenensis, P. stutzeri, P. urumqiensis, P. xanthomarina, P. zhaodongensis | 14 | 73.60–89.69 | 76.39–88.15 |
| P. syringae G | P. amygdali, P. asturieensis, P. avellanae, P. cannabina, P. caricapapayae, P. caspiana, P. cerasi, P. cichorii, P. congelans, P. coronafaciens, P. ficuserectae, P. floridensis, P. meliae, P. ovata, P. savastanoi, P. syringae, P. tremae, P. viridiflava | 18 | 78.36–99.54 | 78.20–94.57 |
| Newly described groups | ||||
| P. pohangensis G | P. mangrovi, P. pohangensis | 2 | 67.02 | 77.07 |
| P. massiliensis G | P. massiliensis, P. typographi | 2 | 80.53 | 76.86 |
| P. rhizosphaerae G | P. baltica, P. coleopterorum, P. rhizosphaerae | 3 | 91.89–94.70 | 88.28–90.55 |
| Orphan groups | ||||
| P. indica G | P. indica | 1 | – | – |
| P. kuykendallii G | P. kuykendallii | 1 | – | – |
| P. thermotolerans G | P. thermotolerans | 1 | – | – |
| P. flexibilis G | P. flexibilis, P. tuomuerensis | 2 | – | – |
| P. fluvialis G | P. fluvialis, P. pharmacofabricae | 2 | – | – |
| P. alcaligenes G | P. alcaligenes | 1 | – | – |
| P. matsuisoli G | P. matsuisoli | 1 | – | – |
| P. turukhanskensis G | P. turukhanskensis | 1 | – | – |
| Groups/Subgroups | Pseudomonas Species | ANIb | Earlier Synonyms | |
|---|---|---|---|---|
| P. aeruginosa group | P. citronellolis | P. humi | 96.70 | P. citronellolis |
| P. nitroreducens | P. nitritireducens | 98.85 | P. nitroreducens | |
| P. oleovorans group | P. oleovorans | P. pseudoalcaligenes | 97.17 | P. oleovorans |
| P. chengduensis | P. sihuiensis | 96.25 | P. chengduensis | |
| P. oryzihabitans group | P. oryzihabitans | P. psychrotolerans | 98.22 | P. oryzihabitans |
| P. luteola | P. zeshuii | 97.87 | P. luteola | |
| P. pertucinogena group | P. phragmitis | P. jilinensis | 98.70 | P. phragmitis |
| P. gallaeciensis | P. abyssi | 97.56 | P. gallaeciensis | |
| P. putida group | P. asiatica | P. pyomelaninifaciens | 99.03 | P. asiatica |
| P. shirazica | 99.17 | |||
| P. stutzeri group | P. chloritidismutans | P. kunmingensis | 96.49 | P. chloritidismutans |
| P. syringae group | P. tremae | P. coronafaciens | 98.74 | P. tremae |
| P. amygdali | P. ficuserectae | 97.42 | P. amygdali | |
| P. meliae | 98.27 | |||
| P. savastanoi | 98.75 | |||
| P. fluorescens group | P. asplenii | P. fuscovaginae | 98.23 | P. asplenii |
| P. veronii | P. panacis | 99.95 | P. veronii | |
| Orphan groups | P. flexibilis | P. tuomuerensis | 98.69 | P. flexibilis |
| P. fluvialis | P. pharmacofabricae | 98.61 | P. fluvialis | |
| Subgroups | Strain | Closest Type Strain | ANIb % | Re-identified Species |
|---|---|---|---|---|
| P. japonica | P. putida CSV86 | P. japonica | 86.94 | Pseudomonas sp. #1 |
| P. reidholzensis | P. putida 02C-26 | P. shirazensis | 97.25 | P. shirazensis |
| P. guariconensis | P. putida IEC33019 | P. guariconensis | 91.47 | Pseudomonas sp. #2 |
| P. putida WP4-W18-CRE-03 | P. guariconensis | 99.38 | P. guariconensis | |
| P. putida WP8-W18-CRE-01 | 99.47 | |||
| P. wayambapalatensis | P. putida NX-1 | P. wayambapalatensis | 94.69 | Pseudomonas sp. #3 |
| P. putida PC2 | 94.78 | |||
| Pseudomonas sp. RW3S2 | P. wayambapalatensis | 99.21 | P. wayambapalatensis | |
| Pseudomonas sp. RW10S2 | 99.26 | |||
| P. farsensis | Pseudomonas sp. SWRI51 | P. farsensis | 98.65 | P. farsensis |
| P. mosselii | Pseudomonas sp. 250J | P. peradeniyensis | 96.15* | Pseudomonas sp. #4 |
| Pseudomonas sp. BW16M2 | P. peradeniyensis | 96.59 | P. peradeniyensis | |
| P. plecoglossicida | P. putida GM84 | P. plecoglossicida | 91.13 | Pseudomonas sp. #5 |
| P. putida GIMC5401-PPKH-115 | P. plecoglossicida | 87.01 | Pseudomonas sp. #6 | |
| P. putida BR-PH17 | P. plecoglossicida | 86.75 | Pseudomonas sp. #7 | |
| P. putida W619 | 85.75 | |||
| P. vlassakiae | P. putida AA7 | P. vlassakiae | 90.88 | Pseudomonas sp. #8 |
| P. putida W5 | P. vlassakiae | 91.64 | Pseudomonas sp. #9 | |
| P. capeferrum | Pseudomonas sp. SWRI68 | P. capeferrum | 98.66 | P. capeferrum |
| Pseudomonas sp. SWRI59 | 98.65 | |||
| P. putida E41 | P. kermanshahensis | 97.48 | P. kermanshahensis | |
| P. putida E46 | 97.55 | |||
| Pseudomonas sp. SWRI50 | 99.39 | |||
| Pseudomonas sp. SWRI67 | 99.99 | |||
| P. putida | P. putida SY153 | P. jutendi | 98.15 | P. jutendi |
| P. putida TIJ-51 | 97.77 | |||
| P. putida GB-1 | P. alloputida | 90.49 | Pseudomonas sp. #10 | |
| P. putida PP112420 | 90.54 | |||
| P. putida S13-1-2 | P. putida | 94.55 | Pseudomonas sp. #11 | |
| P. putida KF715 | 93.73 | Pseudomonas sp. #12 | ||
| P. putida ZXPA-20 | 93.40 | |||
| P. putida H8234 | 93.33 | |||
| P. putida B1 | 93.40 | |||
| P. putida R51 | P. alloputida | 95.00* | Pseudomonas sp. #13 | |
| P. putida BS3701 | P. alloputida | 96.67 | P. alloputida | |
| P. putida MX-2 | 96.49 | |||
| P. putida LS46 | 96.44 | |||
| P. putida 15420352 | 96.42 | |||
| P. putida YC-AE1 | 96.40 | |||
| P. putida T25-27 | 96.51 | |||
| P. monteilii 170620603RE | P. kurunegalensis | 99.45 | P. kurunegalensis | |
| P. monteilii 170918607 | 99.44 | |||
| P. monteilii STW0522-72 | 99.64 | |||
| P. monteilii FDAARGOS171 | 99.77 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Girard, L.; Lood, C.; Höfte, M.; Vandamme, P.; Rokni-Zadeh, H.; van Noort, V.; Lavigne, R.; De Mot, R. The Ever-Expanding Pseudomonas Genus: Description of 43 New Species and Partition of the Pseudomonas putida Group. Microorganisms 2021, 9, 1766. https://doi.org/10.3390/microorganisms9081766
Girard L, Lood C, Höfte M, Vandamme P, Rokni-Zadeh H, van Noort V, Lavigne R, De Mot R. The Ever-Expanding Pseudomonas Genus: Description of 43 New Species and Partition of the Pseudomonas putida Group. Microorganisms. 2021; 9(8):1766. https://doi.org/10.3390/microorganisms9081766
Chicago/Turabian StyleGirard, Léa, Cédric Lood, Monica Höfte, Peter Vandamme, Hassan Rokni-Zadeh, Vera van Noort, Rob Lavigne, and René De Mot. 2021. "The Ever-Expanding Pseudomonas Genus: Description of 43 New Species and Partition of the Pseudomonas putida Group" Microorganisms 9, no. 8: 1766. https://doi.org/10.3390/microorganisms9081766
APA StyleGirard, L., Lood, C., Höfte, M., Vandamme, P., Rokni-Zadeh, H., van Noort, V., Lavigne, R., & De Mot, R. (2021). The Ever-Expanding Pseudomonas Genus: Description of 43 New Species and Partition of the Pseudomonas putida Group. Microorganisms, 9(8), 1766. https://doi.org/10.3390/microorganisms9081766







