Comparative Pan-Genome Analysis of Oral Veillonella Species
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Draft or Complete Genome Sequences, Assemblies and Annotation
2.3. Comparative Pan-Genome Analysis of Oral Veillonella
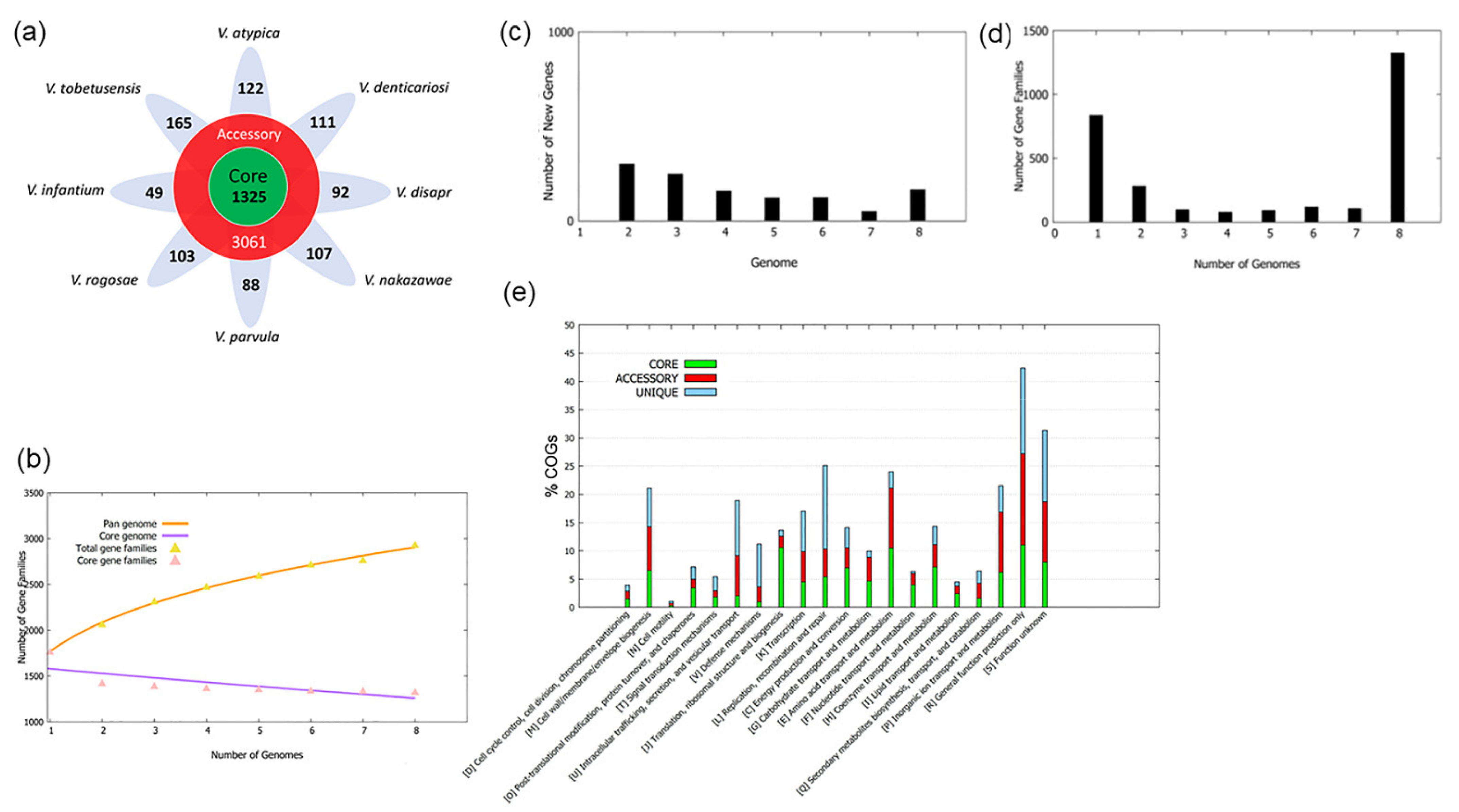
3. Results and Discussion
3.1. Pan-Genome Construction
3.2. Pan-Genomic Analysis
3.3. COG Distribution of Core, Accessory Genome and Unique Genes
3.4. Phylogenetic and Evolutionary Analysis of Oral Veillonella
3.5. KEGG Pathway Mapping of Genes
3.6. Glycolysis and Its Related KEGG Pathways in Carbon Metabolism of Oral Veillonella
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Carlier, J.P. Veillonella. In Bergey’s Manual of Systematics of Archaea and Bacteria; Wiley: Hoboken, NJ, USA, 2015; pp. 1–11. [Google Scholar]
- Parte, A.C. LPSN (List of Prokaryotic Names with Standing Nomenclature) 2021. Available online: www.bacterio.net (accessed on 15 June 2021).
- Kraatz, M.; Taras, D. Veillonella magna sp. nov., isolated from the jejunal mucosa of a healthy pig, and emended description of Veillonella ratti. Int. J. Syst. Evol. Microbiol. 2008, 58, 2755–2761. [Google Scholar] [CrossRef] [PubMed]
- Aujoulat, F.; Bouvet, P.; Jumas-Bilak, E.; Jean-Pierre, H.; Marchandin, H. Veillonella seminalis sp. nov., a novel anaerobic Gram-stain negative coccus from human clinical samples, and emended description of the genus Veillonella. Int. J. Syst. Evol. Microbiol. 2014, 64, 3526–3531. [Google Scholar] [CrossRef] [Green Version]
- Rogosa, M. Anaerobic Gram-negative cocci. In Bergey’s Manual of Systematic Bacteriology; Krieg, N.R., Holt, J.G., Eds.; Williams & Wilkins: Baltimore, MD, USA, 1984; Volume 1, pp. 680–685. [Google Scholar]
- Ng, S.K.C.; Hamilton, I.R. Lactate metabolism by Veillonella parvula. J. Bacteriol. 1971, 105, 999–1005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delwiche, E.A.; Pestka, J.J.; Tortorello, M.L. The veillonellae: Gram-negative cocci with a unique physiology. Annu. Rev. Microbiol. 1985, 39, 175–193. [Google Scholar] [CrossRef]
- Foubert, E.L.; Douglas, H.C. Studies on the anaerobic micrococci. II. The fermentation of lactate by Micrococcus lactilyticus. J. Bacteriol. 1948, 56, 35–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheiman, J.; Luber, J.M.; Chavkin, T.A.; MacDonald, T.; Tung, A.; Pham, L.-D.; Wibowo, M.C.; Wurth, R.C.; Punthambaker, S.; Tierney, B.T.; et al. Metaomics analysis of elite athletes identifies a performance-enhancing microbe that functions via lactate metabolism. Nat. Med. 2019, 25, 1104–1109. [Google Scholar] [CrossRef]
- Mays, T.D.; Holdeman, L.V.; Moore, W.E.C.; Rogosa, M.; Johnson, J.L. Taxonomy of the genus Veillonella Prèvot. Int. J. Syst. Bacteriol. 1982, 32, 28–36. [Google Scholar] [CrossRef]
- Rogosa, M. The genus Veillonella IV. serological groupings, and genus and species emendations. J. Bacteriol. 1965, 90, 704–709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Byun, R.; Carlier, J.P.; Jacques, N.A.; Marchandin, H.; Hunter, N. Veillonella denticariosi sp. nov., isolated from human carious dentine. Int. J. Syst. Evol. Microbiol. 2007, 57, 2844–2848. [Google Scholar] [CrossRef] [PubMed]
- Arif, N.; Do, T.; Byun, R.; Sheehy, E.; Clark, D.; Gilbert, S.C.; Beighton, D. Veillonella rogosae sp. nov., an anaerobic, Gram-negative coccus isolated from dental plaque. Int. J. Syst. Evol. Microbiol. 2008, 58, 581–584. [Google Scholar] [CrossRef]
- Mashima, I.; Kamaguchi, A.; Miyakawa, H.; Nakazawa, F. Veillonella tobetsuensis sp. nov., an anaerobic, Gram-negative coccus isolated from human tongue biofilms. Int. J. Syst. Evol. Microbiol. 2013, 63, 1443–1449. [Google Scholar] [CrossRef] [Green Version]
- Mashima, I.; Liao, Y.C.; Miyakawa, H.; Theodorea, C.F.; Thaweboon, B.; Thaweboom, S.; Scannapieco, F.A.; Nakazawa, F. Veillonella infantium sp. nov., an anaerobic Gram-stain-negative coccus isolated from tongue biofilm of a Thai child. Int. J. Syst. Evol. Microbiol. 2018, 68, 1101–1106. [Google Scholar] [CrossRef] [PubMed]
- Mashima, I.; Theodorea, C.F.; Djais, A.A.; Kunihiro, T.; Kawamura, Y.; Otomo, M.; Saitoh, M.; Tamai, R.; Kiyoura, Y. Veillonella nakazawae sp. nov., an anaerobic Gram-negative coccus isolated from the oral cavity of Japanese children. Int. J. Syst. Evol. Microbiol. 2021, 71. in press. [Google Scholar] [CrossRef]
- Beighton, D.; Clark, D.; Hanakuka, B.; Gilbert, S.; Do, T. The predominant cultivable Veillonella spp. of the tongue of healthy adults identified using rpoB sequencing. Oral Microbiol. Immunol. 2008, 23, 344–347. [Google Scholar] [CrossRef] [PubMed]
- Mashima, I.; Kamaguchi, A.; Nakazawa, F. The distribution and frequency of oral Veillonella spp. in the tongue biofilm of healthy young adults. Curr. Microbiol. 2011, 63, 403–407. [Google Scholar] [CrossRef]
- Mashima, I.; Fujita, M.; Nakatsuka, Y.; Kado, T.; Furuichi, Y.; Sulistyani, H.; Nakazawa, F. The distribution and frequency of Veillonella spp. associated with chronic periodontal diseases. Int. J. Curr. Microbiol. Appl. Sci. 2015, 4, 150–160. [Google Scholar]
- Tanner, A.C.R.; Mathney, J.M.J.; Kent, R.L.; Chalmers, N.I.; Hughes, C.V.; Loo, C.Y.; Pradhan, N.; Kanasi, E.; Hwang, J.; Dahlan, M.A.; et al. Cultivable anaerobic microbiota of severe early childhood caries. J. Clin. Microbiol. 2011, 49, 1464–1474. [Google Scholar] [CrossRef] [Green Version]
- Leuckfeld, I.; Paster, J.B.; Kristoffersen, A.K.; Olsen, I. Diversity of Veillonella spp. from subgingival plaque by polyphasic approach. APMIS 2010, 118, 230–242. [Google Scholar] [CrossRef]
- Takeshita, T.; Matsuo, K.; Furuta, M.; Shibata, Y.; Fukami, K.; Shimazaki, Y.; Akifusa, S.; Han, D.H.; Kim, H.D.; Yokoyama, T.; et al. Distinct composition of the oral indigenous microbiota in South Korean and Japanese adults. Sci. Rep. 2014, 4, 6990. [Google Scholar] [CrossRef] [Green Version]
- Jiang, S.; Gao, X.; Jin, L.; Lo, E.C.M. Salivary microbiome diversity in caries-free and caries-affected children. Int. J. Mol. Sci. 2016, 17, 1978. [Google Scholar] [CrossRef] [Green Version]
- Jesus, V.C.D.; Shikder, R.; Oryniak, D.; Mann, K.; Alamri, A.; Mittermuller, B.; Duan, K.; Hu, P.; Schroth, R.J.; Chelikani, P. Sex-based diverse plaque microbiota in children with severe caries. J. Dent. Res. 2020, 99, 703–712. [Google Scholar] [CrossRef]
- Mashima, I.; Theodorea, C.F.; Thaweboon, B.; Thaweboon, S.; Scannapieco, F.A.; Nakazawa, F. Exploring the salivary microbiome of children stratified by the oral hygiene index. PLoS ONE 2017, 12, e0185274. [Google Scholar] [CrossRef]
- Kolenbrander, P.E. The genus Veillonella. In The Prokaryotes volume 4 Bacteria, Firmicutes, Cyanobacteria; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2006; pp. 1022–1040. [Google Scholar]
- Periasamy, S.; Kolenbrander, P.E. Central role of the early colonizer Veillonella sp. in establishing multispecies biofilm communities with initial, middle, and late colonizers of enamel. J. Bacteriol. 2010, 192, 2965–2972. [Google Scholar] [CrossRef] [Green Version]
- Mashima, I.; Nakazawa, F. The influence of oral Veillonella species on biofilms formed by Streptococcus species. Anaerobe 2014, 28, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Mashima, I.; Nakazawa, F. Interaction between Streptococcus spp. and Veillonella tobetsuensis in the early stages of oral biofilm formation. J. Bacteriol. 2015, 197, 2104–2111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knapp, S.; Brodal, C.; Peterson, J.; Qi, F.; Kreth, J.; Merritt, J. Natural competence is common among clinical isolates of Veillonella parvula and is useful for genetic manupilation of this key member of the oral microbiome. Front. Cell. Infect. Microbiol. 2017, 7, 139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, M.; Liu, G.; Chen, Y.; Wang, D.; Zhang, Y. Comparative genomics uncovers the genetic diversity and characters of Veillonella atypica and provides insights into its potential applications. Front. Microbiol. 2020, 11, 1219. [Google Scholar] [CrossRef]
- Poppleton, D.I.; Duchateau, M.; Hourdel, V.; Matondo, M.; Flechsler, J.; Klingl, A.; Beloin, C.; Gribaldo, S. Outer membrane proteome of Veillonella parvula: A diderm firmicute of the human microbiome. Front. Microbiol. 2017, 8, 1215. [Google Scholar] [CrossRef]
- Chaudhari, N.M.; Gupta, V.K.; Dutta, C. BPGA- an ultra-fast pan-genome analysis pipeline. Sci. Rep. 2016, 6, 24373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ochman, H.; Lerat, E.; Daubin, V. Examining bacterial species under the specter of gene transfer and exchange. Proc. Natl. Acad. Sci. USA 2005, 102, 6595–6599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Read, T.D.; Ussery, D.W. Opening the pan-genomics box. Curr. Op. Microbiol. 2006, 9, 496–498. [Google Scholar] [CrossRef]
- Tettelin, H.; Masignani, H.; Cieslewicz, M.J.; Donati, C.; Medini, D.; Ward, N.L.; Angiuoli, S.V.; Crabtree, J.; Jones, A.L.; Durkin, A.S.; et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. USA 2005, 102, 13950–13955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sambrook, J.; Russell, D.W. Purification of nucleic acids by extraction with phenol:chloroform. Cold Spring Harb. Protoc. 2006, pdb.prot4455. [Google Scholar] [CrossRef]
- Mashima, I.; Liao, Y.C.; Sabharwal, A.; Haase, E.M.; Nakazawa, F.; Scannapieco, F.A. Draft genome sequences of four strains of recently established novel Veillonella species isolated from human oral cavities. Genome Announc. 2018, 12, e00259-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, Y.C.; Lin, H.H.; Sabharwal, A.; Haase, E.M.; Scannapieco, F.A. MyPro: A seamless pipeline for automated prokaryotic genome assembly and annotation. J. Microbial. Meth. 2015, 113, 72–74. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Watanabe, M.I.; Tanabe, M. KEGG: Integrating virus and cellular organisms. Nuc. Acid Res. 2021, 49, D545–D551. [Google Scholar] [CrossRef] [PubMed]
- Guangchuang, Y.; Wang, L.G.; Han, Y.; Yu, Q.H. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics 2012, 16, 284–287. [Google Scholar]
- Darling, A.E.; Mau, B.; Perna, N.T. ProgressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef] [Green Version]
- Veras, A.; Araujo, F.; Pinheiro, K.; Guimarães, L.; Azevedo, V.; Soares, S.; da Silva, A.D.C.; Ramos, R. Pan4Draft: A computational tool to improve the accuracy of pan-genomic analysis using draft genomes. Sci. Rep. 2018, 8, 9670. [Google Scholar] [CrossRef]
- Bravo, C.; Martinez, V. Whole-genome comparative analysis of the pathogen Piscirickettsia salmonis. Vet. Microbiol. 2016, 196, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Livingstone, P.G.; Morphew, R.M.; Whitworth, D.E. Genome sequencing and pan-genome analysis of 23 Corallococcus spp. strains reveal unexpected diversity, with particular plasticity of predatory gene sets. Front. Microbiol. 2018, 9, 3187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mashima, I.; Nakazawa, F. Draft genome sequence of Veillonella tobetsuensis ATCC BAA-2400T isolated from human tongue biofilm. Genome Announc. 2015, 20, e00808-15. [Google Scholar]
- Mashima, I.; Nakazawa, F.; Tamai, R.; Kiyoura, Y. Complete genome sequence of Veillonella nakazawae JCM 33966T (= CCUG 74597T), isolated from the oral cavity of Japanese children. Microbiol. Resourc. Announc. 2021, 10, e00279-21. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Kiryutin, B.; Koonin, E.V.; Krylov, D.M.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; et al. The COG database: An updated version includes eukaryotes. BMC Bioinform. 2003, 11, 4–41. [Google Scholar]
- Paradis, E.; Schliep, K. Ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 2019, 35, 526–528. [Google Scholar] [CrossRef] [PubMed]
- Valentine, R.C.; Jackson, R.L.; Wolfe, R.S. Role of ferredoxin in hydrogen metabolism of Micrococcus lactilyticus. Biochem. Biophys. Res. Commun. 1962, 7, 453–456. [Google Scholar] [CrossRef]
- Ng, S.K.C.; Hamilton, I.R. Purification and regulatory properties of pyruvate kinase from Veillonella parvula. J. Bacteriol. 1975, 122, 1274–1282. [Google Scholar] [CrossRef] [Green Version]




| Genome No. | Species Name | Strain | Type Strain | Assembly Level | Genome Size (bp) | N50 | G+C (%) | Number of Genes | Number of CDSs | Number of Proteins | Data Source of Nucleotide Sequence | Accession Numbers |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Veillonella atypica | ATCC 17744 | YES | Draft | 2,037,410 | 300,566 | 39.0 | 1928 | 1864 | 1832 | https://ftp.ncbi.nlm.nih.gov/genomes/all/GCA/002/959/915/GCA_002959915.1_ASM295991v1 (accessed on 22nd July 2021) | PPDE01000000 |
| 2 | Veillonella denticariosi | JCM 15641 | YES | Draft | 1,981,866 | 600,371 | 42.9 | 1852 | 1783 | 1746 | https://ftp.ncbi.nlm.nih.gov/genomes/all/GCA/002/959/855/GCA_002959855.1_ASM295985v1 (accessed on 22nd July 2021) | PPDB00000000 |
| 3 | Veillonella dispar | ATCC 17748 | YES | Draft | 2,116,567 | 498,249 | 38.9 | 1991 | 1926 | 1903 | https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/160/015/GCF_000160015.1_ASM16001v1/ (accessed on 5th September 2017) | NZ_ACIK00000000 |
| 4 | Veillonella nakazawae | JCM 33966 | YES | Complete | 2,097,818 | 2,097,818 | 38.6 | 1957 | 1893 | 1925 | https://ftp.ncbi.nlm.nih.gov/genomes/all/GCA/013/393/365/GCA_013393365.1_ASM1339336v1/ (accessed on 8th July 2020) | AP022321 |
| 5 | Veillonella parvula | DSM 2008 | YES | Complete | 2,132,142 | 2,132,142 | 38.6 | 1904 | 1840 | 1824 | https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/024/945/GCF_000024945.1_ASM2494v1/ (accessed on 5th September 2017) | NC_013520.1 |
| 6 | Veillonella rogosae | JCM 15642 | YES | Draft | 2,187,106 | 175,154 | 38.9 | 2068 | 2002 | 1951 | https://ftp.ncbi.nlm.nih.gov/genomes/all/GCA/002/959/775/GCA_002959775.1_ASM295977v1 (accessed on 22nd July 2021) | PPCX00000000 |
| 7 | Veillonella infantium | JCM 31738 | YES | Draft | 2,021,343 | 235,046 | 36.6 | 1899 | 1837 | 1809 | https://ftp.ncbi.nlm.nih.gov/genomes/all/GCA/002/959/895/GCA_002959895.1_ASM295989v1 (accessed on 22nd July 2021) | PPDD00000000 |
| 8 | Veillonella tobetuensis | ATCC BAA-2400 | YES | Draft | 2,161,277 | 225,588 | 38.5 | 2018 | 1948 | 1896 | https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/001/078/375/GCF_001078375.1_ASM107837v1/ (accessed on 5th September 2017) | NZ_BBXI00000000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mashima, I.; Liao, Y.-C.; Lin, C.-H.; Nakazawa, F.; Haase, E.M.; Kiyoura, Y.; Scannapieco, F.A. Comparative Pan-Genome Analysis of Oral Veillonella Species. Microorganisms 2021, 9, 1775. https://doi.org/10.3390/microorganisms9081775
Mashima I, Liao Y-C, Lin C-H, Nakazawa F, Haase EM, Kiyoura Y, Scannapieco FA. Comparative Pan-Genome Analysis of Oral Veillonella Species. Microorganisms. 2021; 9(8):1775. https://doi.org/10.3390/microorganisms9081775
Chicago/Turabian StyleMashima, Izumi, Yu-Chieh Liao, Chieh-Hua Lin, Futoshi Nakazawa, Elaine M. Haase, Yusuke Kiyoura, and Frank A. Scannapieco. 2021. "Comparative Pan-Genome Analysis of Oral Veillonella Species" Microorganisms 9, no. 8: 1775. https://doi.org/10.3390/microorganisms9081775
APA StyleMashima, I., Liao, Y.-C., Lin, C.-H., Nakazawa, F., Haase, E. M., Kiyoura, Y., & Scannapieco, F. A. (2021). Comparative Pan-Genome Analysis of Oral Veillonella Species. Microorganisms, 9(8), 1775. https://doi.org/10.3390/microorganisms9081775







