Microbiome Analysis of Carious Lesions in Pre-School Children with Early Childhood Caries and Congenital Heart Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Dental Examination and Sampling
2.2. DNA Extraction and 16S RNA Gene Amplicon Sequencing
2.3. Statistical Analysis/Bioinformatics
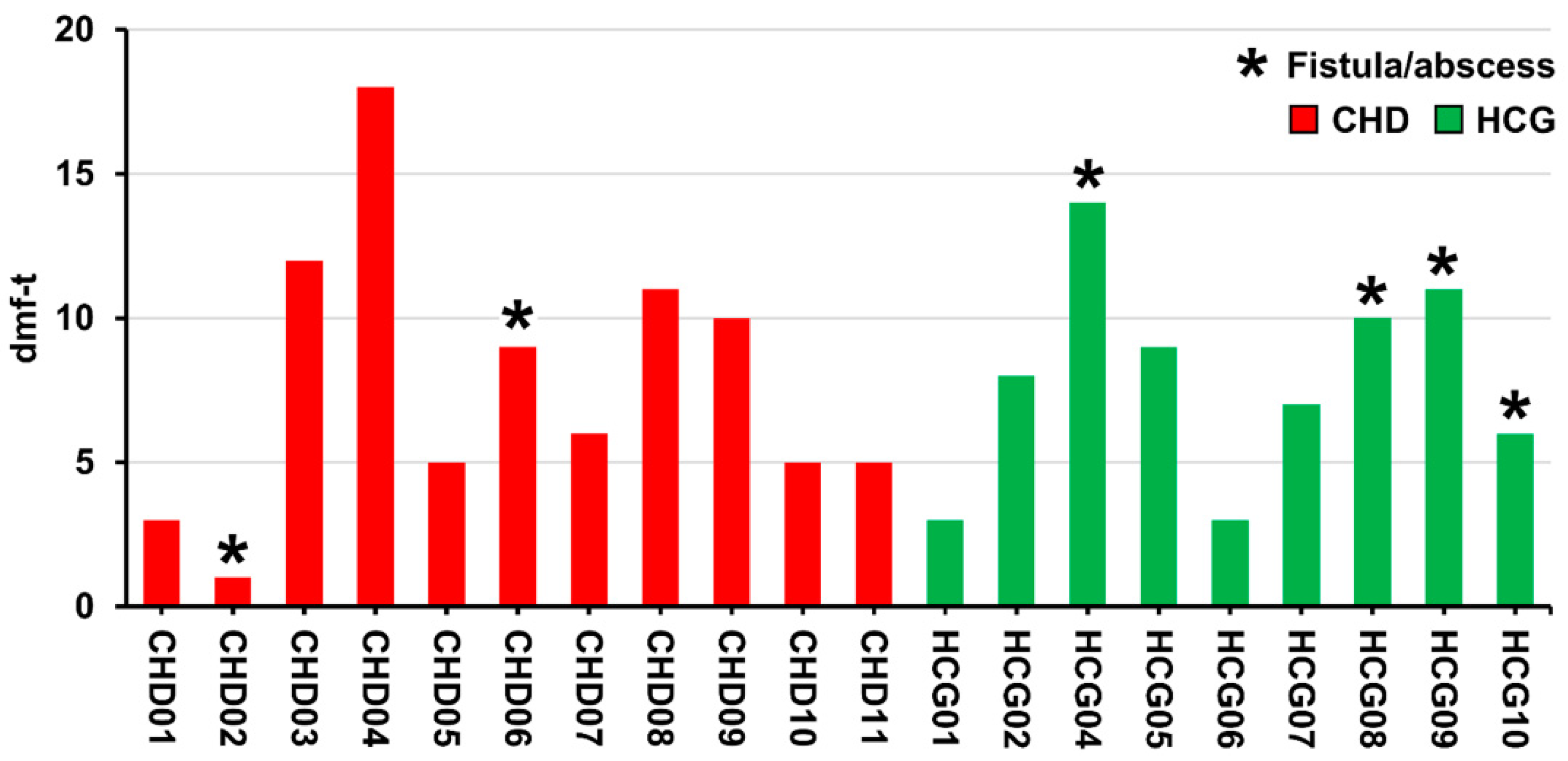
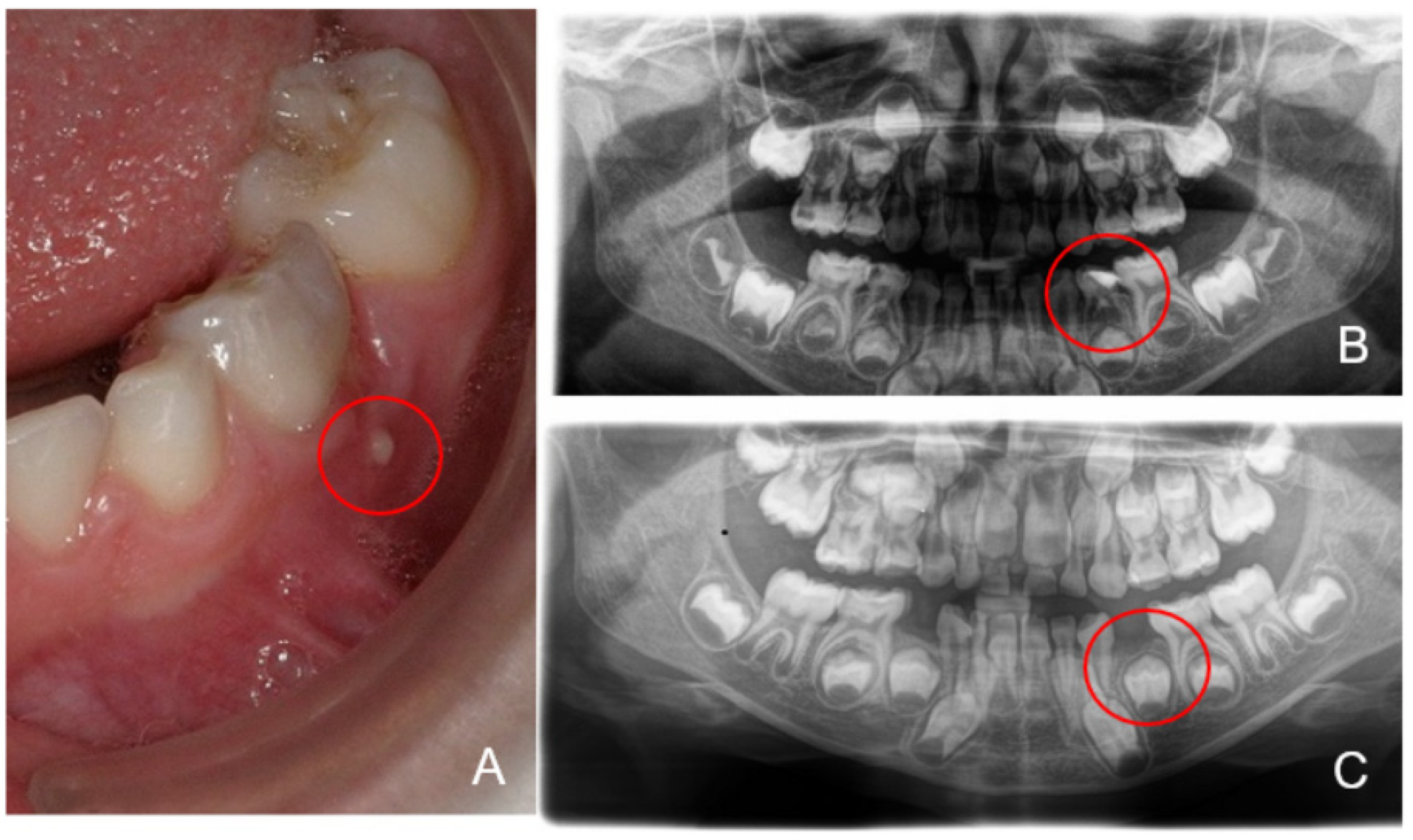
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tinanoff, N.; Baez, R.J.; Diaz Guillory, C.; Donly, K.J.; Feldens, C.A.; McGrath, C.; Phantumvanit, P.; Pitts, N.B.; Seow, W.K.; Sharkov, N.; et al. Early childhood caries epidemiology, aetiology, risk assessment, societal burden, management, education, and policy: Global perspective. Int. J. Paediatr. Dent. 2019, 29, 238–248. [Google Scholar] [CrossRef]
- Albadri, S.S.; Jarad, F.D.; Lee, G.T.; Mackie, I.C. The frequency of repeat general anaesthesia for teeth extractions in children. Int. J. Paediatr. Dent. 2006, 16, 45–48. [Google Scholar] [CrossRef]
- Schulz-Weidner, N.; Logeswaran, T.; Jux, C.; Schlenz, M.A.; Kramer, N.; Bulski, J.C. Evaluation of the effectiveness of an interdisciplinary preventive oral hygiene program for children with congenital heart disease. Int. J. Environ. Res. Public Health 2021, 18, 3497. [Google Scholar] [CrossRef]
- Fakhruddin, K.S.; Ngo, H.C.; Samaranayake, L.P. Cariogenic microbiome and microbiota of the early primary dentition: A contemporary overview. Oral Dis. 2019, 25, 982–995. [Google Scholar] [CrossRef]
- Becker, M.R.; Paster, B.J.; Leys, E.J.; Moeschberger, M.L.; Kenyon, S.G.; Galvin, J.L.; Boches, S.K.; Dewhirst, F.E.; Griffen, A.L. Molecular analysis of bacterial species associated with childhood caries. J. Clin. Microbiol. 2002, 40, 1001–1009. [Google Scholar] [CrossRef]
- Tanner, A.C.; Kent, R.L., Jr.; Holgerson, P.L.; Hughes, C.V.; Loo, C.Y.; Kanasi, E.; Chalmers, N.I.; Johansson, I. Microbiota of severe early childhood caries before and after therapy. J. Dent. Res. 2011, 90, 1298–1305. [Google Scholar] [CrossRef] [PubMed]
- Finucane, D. Rationale for restoration of carious primary teeth: A review. Eur. Arch. Paediatr. Dent. 2012, 13, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Horiuchi, M.; Washio, J.; Mayanagi, H.; Takahashi, N. Transient acid-impairment of growth ability of oral Streptococcus, Actinomyces and Lactobacillus: A possible ecological determinant in dental plaque. Oral Microbiol. Immunol. 2009, 24, 319–324. [Google Scholar] [CrossRef]
- Richards, V.P.; Alvarez, A.J.; Luce, A.R.; Bedenbaugh, M.; Mitchell, M.L.; Burne, R.A.; Nascimento, M.M. Microbiomes of site-specific dental plaques from children with different caries status. Infect. Immun. 2017, 85. [Google Scholar] [CrossRef]
- Yamashita, Y.; Takeshita, T. The oral microbiome and human health. J. Oral Sci. 2017, 59, 201–206. [Google Scholar] [CrossRef]
- Xin, X.; Junzhi, H.; Xuedong, Z. Oral microbiota: A promising predictor of human oral and systemic diseases. Hua Xi Kou Qiang Yi Xue Za Zhi 2015, 33, 555–560. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, X.; Li, H.; Ni, C.; Du, Z.; Yan, F. Human oral microbiota and its modulation for oral health. Biomed. Pharmacother. 2018, 99, 883–893. [Google Scholar] [CrossRef]
- Sampaio-Maia, B.; Caldas, I.M.; Pereira, M.L.; Pérez-Mongiovi, D.; Araujo, R. The oral microbiome in health and its implication in oral and systemic diseases. Adv. Appl. Microbiol. 2016, 97, 171–210. [Google Scholar] [CrossRef]
- Xiao, J.; Fiscella, K.A.; Gill, S.R. Oral microbiome: Possible harbinger for children’s health. Int. J. Oral Sci. 2020, 12, 12. [Google Scholar] [CrossRef]
- Griessl, T.; Zechel-Gran, S.; Olejniczak, S.; Weigel, M.; Hain, T.; Domann, E. High-resolution taxonomic examination of the oral microbiome after oil pulling with standardized sunflower seed oil and healthy participants: A pilot study. Clin. Oral Investig. 2020. [Google Scholar] [CrossRef]
- Hirschfeld, J.; Kawai, T. Oral inflammation and bacteremia: Implications for chronic and acute systemic diseases involving major organs. Cardiovasc. Haematol. Disord. Drug Targets 2015, 15, 70–84. [Google Scholar] [CrossRef]
- Lockhart, P.B.; Brennan, M.T.; Thornhill, M.; Michalowicz, B.S.; Noll, J.; Bahrani-Mougeot, F.K.; Sasser, H.C. Poor oral hygiene as a risk factor for infective endocarditis-related bacteremia. J. Am. Dent. Assoc. 2009, 140, 1238–1244. [Google Scholar] [CrossRef]
- Berbari, E.F.; Cockerill, F.R., III; Steckelberg, J.M. Infective endocarditis due to unusual or fastidious microorganisms. Mayo Clin. Proc. 1997, 72, 532–542. [Google Scholar] [CrossRef]
- Hughes, S.; Balmer, R.; Moffat, M.; Willcoxson, F. The dental management of children with congenital heart disease following the publication of paediatric congenital heart disease standards and specifications. Br. Dent. J. 2019, 226, 447–452. [Google Scholar] [CrossRef]
- Slocum, C.; Kramer, C.; Genco, C.A. Immune dysregulation mediated by the oral microbiome: Potential link to chronic inflammation and atherosclerosis. J. Intern. Med. 2016, 280, 114–128. [Google Scholar] [CrossRef] [PubMed]
- Hallett, K.B.; Radford, D.J.; Seow, W.K. Oral health of children with congenital cardiac diseases: A controlled study. Pediatric Dent. 1992, 14, 224–230. [Google Scholar]
- Duval, X.; Delahaye, F.; Alla, F.; Tattevin, P.; Obadia, J.F.; Le Moing, V.; Doco-Lecompte, T.; Celard, M.; Poyart, C.; Strady, C.; et al. Temporal trends in infective endocarditis in the context of prophylaxis guideline modifications: Three successive population-based surveys. J. Am. Coll. Cardiol. 2012, 59, 1968–1976. [Google Scholar] [CrossRef] [PubMed]
- Demmer, R.T.; Desvarieux, M. Periodontal infections and cardiovascular disease: The heart of the matter. J. Am. Dent. Assoc. 2006, 137 (Suppl. 2), 14S–20S, quiz 38S. [Google Scholar] [CrossRef][Green Version]
- Wang, Y.; Zeng, X.; Yang, X.; Que, J.; Du, Q.; Zhang, Q.; Zou, J. Oral health, caries risk profiles, and oral microbiome of pediatric patients with leukemia submitted to chemotherapy. Biomed. Res. Int. 2021, 2021, 6637503. [Google Scholar] [CrossRef]
- Warnes, C.A.; Liberthson, R.; Danielson, G.K.; Dore, A.; Harris, L.; Hoffman, J.I.; Somerville, J.; Williams, R.G.; Webb, G.D. Task force 1: The changing profile of congenital heart disease in adult life. J. Am. Coll. Cardiol. 2001, 37, 1170–1175. [Google Scholar] [CrossRef]
- Uebereck, C.; Kühnisch, J.; Michel, R.; Taschner, M.; Frankenberger, R.; Krämer, N. Zahngesundheit bayrischer schulkinder 2015/16. Oralprophylaxe Kinderzahnheilkd 2017, 39, 161–171. [Google Scholar] [CrossRef]
- Elyassi Gorji, N.; Nasiri, P.; Malekzadeh Shafaroudi, A.; Moosazadeh, M. Comparison of dental caries (DMFT and DMFS indices) between asthmatic patients and control group in Iran: A meta-analysis. Asthma Res. Pract. 2021, 7, 2. [Google Scholar] [CrossRef] [PubMed]
- Goswami, M.; Rajwar, A.S. Evaluation of cavitated and non-cavitated carious lesions using the WHO basic methods, ICDAS-II and laser fluorescence measurements. J. Indian Soc. Pedod. Prev. Dent. 2015, 33, 10–14. [Google Scholar] [CrossRef]
- Pitts, N.; Baez, R.; Diaz-Guallory, C.; Donly, K.; Feldens, C.; McGrath, C.; Phantumvanit, P.; Seow, K.; Sharkov, N.; Tinanoff, N.; et al. Early childhood caries: IAPD bangkok declaration. Int. J. Paediatr. Dent. 2019, 29, 384–386. [Google Scholar] [CrossRef]
- American Academy of Pediatric Dentistry. Policy on early childhood caries (ECC): Classifications, consequences, and preventive strategies. Pediatric Dent. 2016, 38, 52–54. [Google Scholar]
- DAJ. Grundsätze für Maßnahmen zur Förderung der Mundgesundheit im Rahmen der Gruppenprophylaxe nach § 21 SGB V. Available online: https://www.daj.de/fileadmin/user_upload/PDF_Downloads/grundsaetze.pdf (accessed on 16 August 2021).
- Dabrowski, A.N.; Shrivastav, A.; Conrad, C.; Komma, K.; Weigel, M.; Dietert, K.; Gruber, A.D.; Bertrams, W.; Wilhelm, J.; Schmeck, B.; et al. Peptidoglycan recognition protein 4 limits bacterial clearance and inflammation in lungs by control of the gut microbiota. Front. Immunol. 2019, 10, 2106. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 1), 4516–4522. [Google Scholar] [CrossRef]
- Regier, A.A.; Farjoun, Y.; Larson, D.E.; Krasheninina, O.; Kang, H.M.; Howrigan, D.P.; Chen, B.J.; Kher, M.; Banks, E.; Ames, D.C.; et al. Functional equivalence of genome sequencing analysis pipelines enables harmonized variant calling across human genetics projects. Nat. Commun. 2018, 9, 4038. [Google Scholar] [CrossRef] [PubMed]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahe, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- NCBI RefSeq Targeted Loci Project. NCBI RefSeq Targeted Loci Project. Available online: https://www.ncbi.nlm.nih.gov/refseq/targetedloci/ (accessed on 16 August 2021).
- Henne, K.; Rheinberg, A.; Melzer-Krick, B.; Conrads, G. Aciduric microbial taxa including Scardovia wiggsiae and Bifidobacterium spp. in caries and caries free subjects. Anaerobe 2015, 35, 60–65. [Google Scholar] [CrossRef]
- Kalpana, B.; Prabhu, P.; Bhat, A.H.; Senthilkumar, A.; Arun, R.P.; Asokan, S.; Gunthe, S.S.; Verma, R.S. Bacterial diversity and functional analysis of severe early childhood caries and recurrence in India. Sci. Rep. 2020, 10, 21248. [Google Scholar] [CrossRef]
- Hurley, E.; Barrett, M.P.J.; Kinirons, M.; Whelton, H.; Ryan, C.A.; Stanton, C.; Harris, H.M.B.; O’Toole, P.W. Comparison of the salivary and dentinal microbiome of children with severe-early childhood caries to the salivary microbiome of caries-free children. BMC Oral Health 2019, 19, 13. [Google Scholar] [CrossRef]
- Liu, G.; Wu, C.; Abrams, W.R.; Li, Y. Structural and functional characteristics of the microbiome in deep-dentin caries. J. Dent. Res. 2020, 99, 713–720. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.L.; Siqueira, J.F., Jr.; Rôças, I.N.; Jesus, E.C.; Rosado, A.S.; Tiedje, J.M. Comparing the bacterial diversity of acute and chronic dental root canal infections. PLoS ONE 2011, 6, e28088. [Google Scholar] [CrossRef]
- Nardello, L.C.L.; Amado, P.P.P.; Franco, D.C.; Cazares, R.X.R.; Nogales, C.G.; Mayer, M.P.A.; Karygianni, L.; Thurnheer, T.; Pinheiro, E.T. Next-Generation sequencing to assess potentially active bacteria in endodontic infections. J. Endod. 2020, 46, 1105–1112. [Google Scholar] [CrossRef] [PubMed]
- Obata, M.; Ohtsuji, M.; Iida, Y.; Shirai, T.; Hirose, S.; Nishimura, H. Genome-wide genetic study in autoimmune disease-prone mice. Methods Mol. Biol. 2014, 1142, 111–141. [Google Scholar] [CrossRef]
- Dashper, S.G.; Mitchell, H.L.; KA, L.C.; Carpenter, L.; Gussy, M.G.; Calache, H.; Gladman, S.L.; Bulach, D.M.; Hoffmann, B.; Catmull, D.V.; et al. Temporal development of the oral microbiome and prediction of early childhood caries. Sci. Rep. 2019, 9, 19732. [Google Scholar] [CrossRef]
- Feldens, C.A.; Rodrigues, P.H.; de Anastacio, G.; Vitolo, M.R.; Chaffee, B.W. Feeding frequency in infancy and dental caries in childhood: A prospective cohort study. Int. Dent. J. 2018, 68, 113–121. [Google Scholar] [CrossRef]
- Ghazal, T.S.; Levy, S.M.; Childers, N.K.; Carter, K.D.; Caplan, D.J.; Warren, J.J.; Cavanaugh, J.E.; Kolker, J. Mutans streptococci and dental caries: A new statistical modeling approach. Caries Res. 2018, 52, 246–252. [Google Scholar] [CrossRef]
- Marchant, S.; Brailsford, S.R.; Twomey, A.C.; Roberts, G.J.; Beighton, D. The predominant microflora of nursing caries lesions. Caries Res. 2001, 35, 397–406. [Google Scholar] [CrossRef]
- Hong, B.Y.; Lee, T.K.; Lim, S.M.; Chang, S.W.; Park, J.; Han, S.H.; Zhu, Q.; Safavi, K.E.; Fouad, A.F.; Kum, K.Y. Microbial analysis in primary and persistent endodontic infections by using pyrosequencing. J. Endod. 2013, 39, 1136–1140. [Google Scholar] [CrossRef]
- Chalmers, N.I.; Palmer, R.J., Jr.; Cisar, J.O.; Kolenbrander, P.E. Characterization of a Streptococcus sp.-Veillonella sp. community micromanipulated from dental plaque. J. Bacteriol. 2008, 190, 8145–8154. [Google Scholar] [CrossRef]
- Macy, J.M.; Yu, I.; Caldwell, C.; Hungate, R.E. Reliable sampling method for analysis of the ecology of the human alimentary tract. Appl. Environ. Microbiol. 1978, 35, 113–120. [Google Scholar] [CrossRef]
- Ohara-Nemoto, Y.; Kishi, K.; Satho, M.; Tajika, S.; Sasaki, M.; Namioka, A.; Kimura, S. Infective endocarditis caused by Granulicatella elegans originating in the oral cavity. J. Clin. Microbiol. 2005, 43, 1405–1407. [Google Scholar] [CrossRef]
- Clayton, J.J.; Baig, W.; Reynolds, G.W.; Sandoe, J.A.T. Endocarditis caused by Propionibacterium species: A report of three cases and a review of clinical features and diagnostic difficulties. J. Med. Microbiol. 2006, 55, 981–987. [Google Scholar] [CrossRef]
- Koren, O.; Spor, A.; Felin, J.; Fak, F.; Stombaugh, J.; Tremaroli, V.; Behre, C.J.; Knight, R.; Fagerberg, B.; Ley, R.E.; et al. Human oral, gut and plaque microbiota in patients with atherosclerosis. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 1), 4592–4598. [Google Scholar] [CrossRef] [PubMed]
- Hossain, H.; Ansari, F.; Schulz-Weidner, N.; Wetzel, W.E.; Chakraborty, T.; Domann, E. Clonal identity of Candida albicans in the oral cavity and the gastrointestinal tract of pre-school children. Oral Microbiol. Immunol. 2003, 18, 302–308. [Google Scholar] [CrossRef]
- Graves, D.T.; Corrêa, J.D.; Silva, T.A. The oral microbiota is modified by systemic diseases. J. Dent. Res. 2019, 98, 148–156. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schulz-Weidner, N.; Weigel, M.; Turujlija, F.; Komma, K.; Mengel, J.P.; Schlenz, M.A.; Bulski, J.C.; Krämer, N.; Hain, T. Microbiome Analysis of Carious Lesions in Pre-School Children with Early Childhood Caries and Congenital Heart Disease. Microorganisms 2021, 9, 1904. https://doi.org/10.3390/microorganisms9091904
Schulz-Weidner N, Weigel M, Turujlija F, Komma K, Mengel JP, Schlenz MA, Bulski JC, Krämer N, Hain T. Microbiome Analysis of Carious Lesions in Pre-School Children with Early Childhood Caries and Congenital Heart Disease. Microorganisms. 2021; 9(9):1904. https://doi.org/10.3390/microorganisms9091904
Chicago/Turabian StyleSchulz-Weidner, Nelly, Markus Weigel, Filip Turujlija, Kassandra Komma, Jan Philipp Mengel, Maximiliane Amelie Schlenz, Julia Camilla Bulski, Norbert Krämer, and Torsten Hain. 2021. "Microbiome Analysis of Carious Lesions in Pre-School Children with Early Childhood Caries and Congenital Heart Disease" Microorganisms 9, no. 9: 1904. https://doi.org/10.3390/microorganisms9091904
APA StyleSchulz-Weidner, N., Weigel, M., Turujlija, F., Komma, K., Mengel, J. P., Schlenz, M. A., Bulski, J. C., Krämer, N., & Hain, T. (2021). Microbiome Analysis of Carious Lesions in Pre-School Children with Early Childhood Caries and Congenital Heart Disease. Microorganisms, 9(9), 1904. https://doi.org/10.3390/microorganisms9091904






