Valorization Potential of a Novel Bacterial Strain, Bacillus altitudinis RSP75, towards Lignocellulose Bioconversion: An Assessment of Symbiotic Bacteria from the Stored Grain Pest, Tribolium castaneum
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents and Substrates
2.2. Identification and Dissection of Insects
2.3. Isolation and Screening for Cellulolytic Bacteria
2.4. Biotyping by MALDI-TOF
2.5. Identification and Phylogenetic Analysis
2.6. Biochemical Characterization
2.7. Effect of pH and Temperature on Cellulase Activity
2.8. Growth Curve
2.9. Enzyme Assays
2.10. Determination of Substrate Degradation Ratio
2.11. Electron Microscopy and Characterization of Hydrolyzed Substrate
2.12. GC-MS Analysis
2.13. Statistical Analyses
3. Results
3.1. Identification of the Insect
3.2. Isolation and Identification of Bacteria by MALDI-TOF
3.3. Screening and Selection of the Potential Bacteria
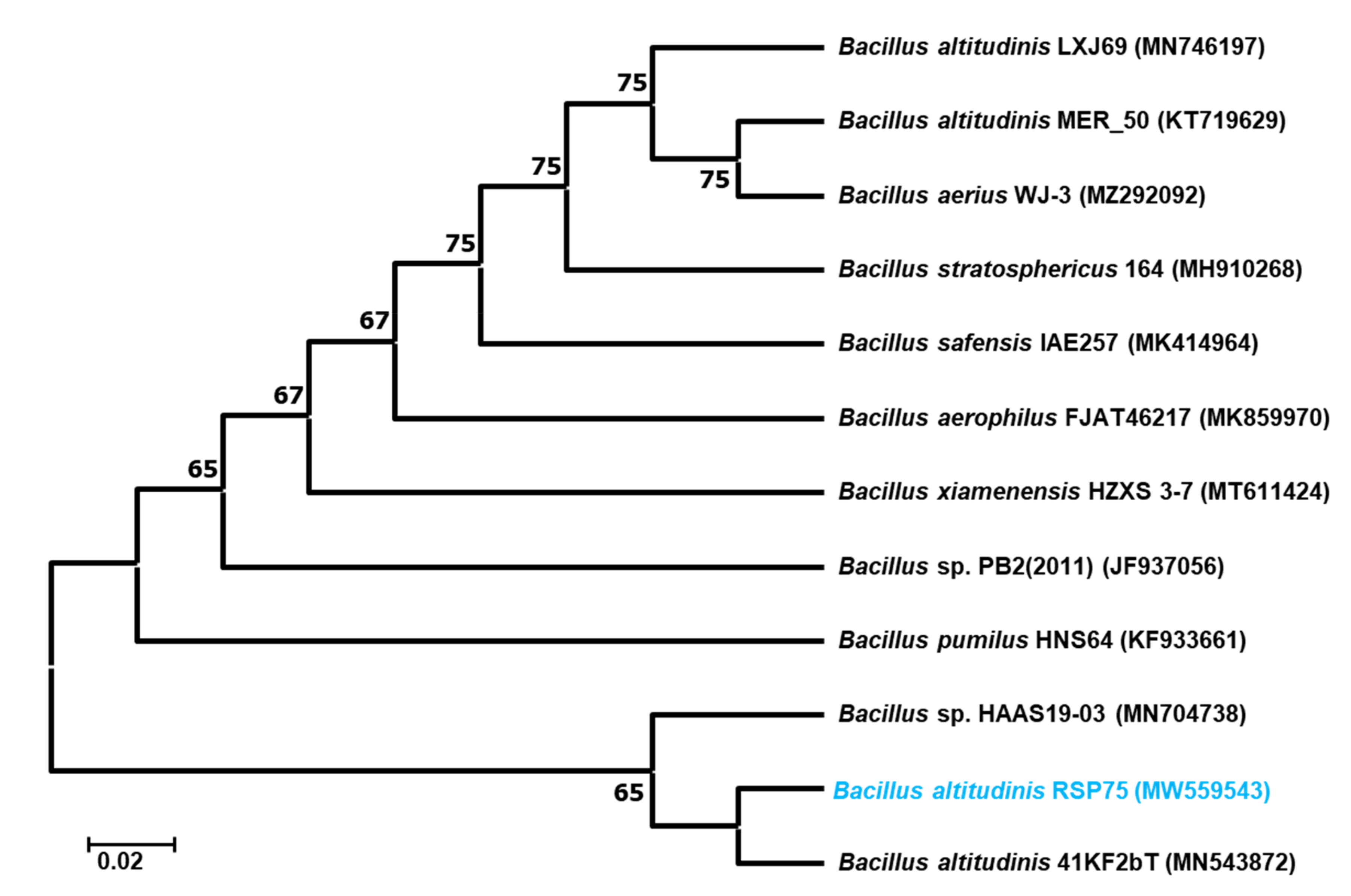
3.4. Molecular Identification and Phylogenetic Analysis of the Bacteria
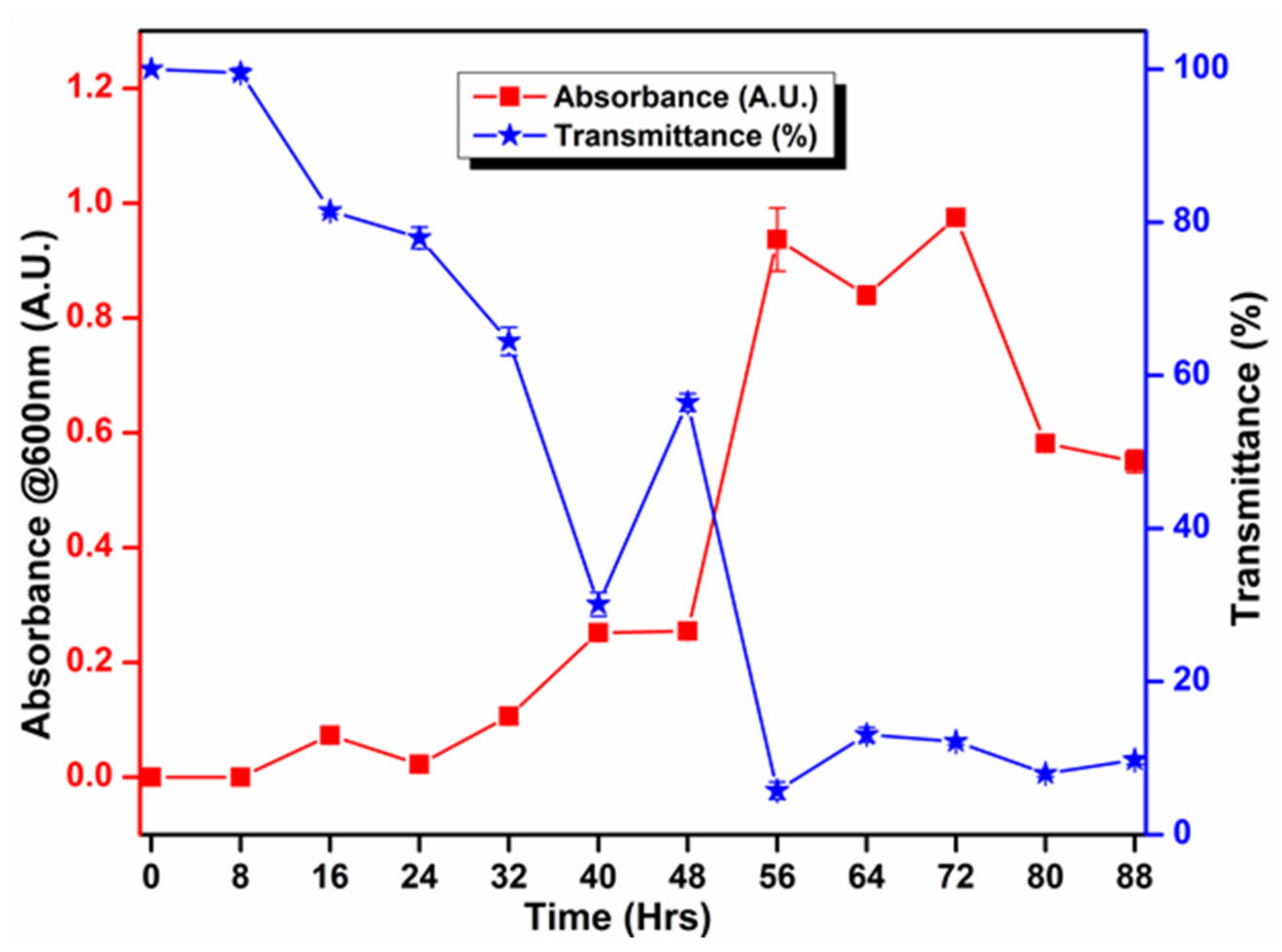
3.5. Biochemical and Growth Curve Analyses
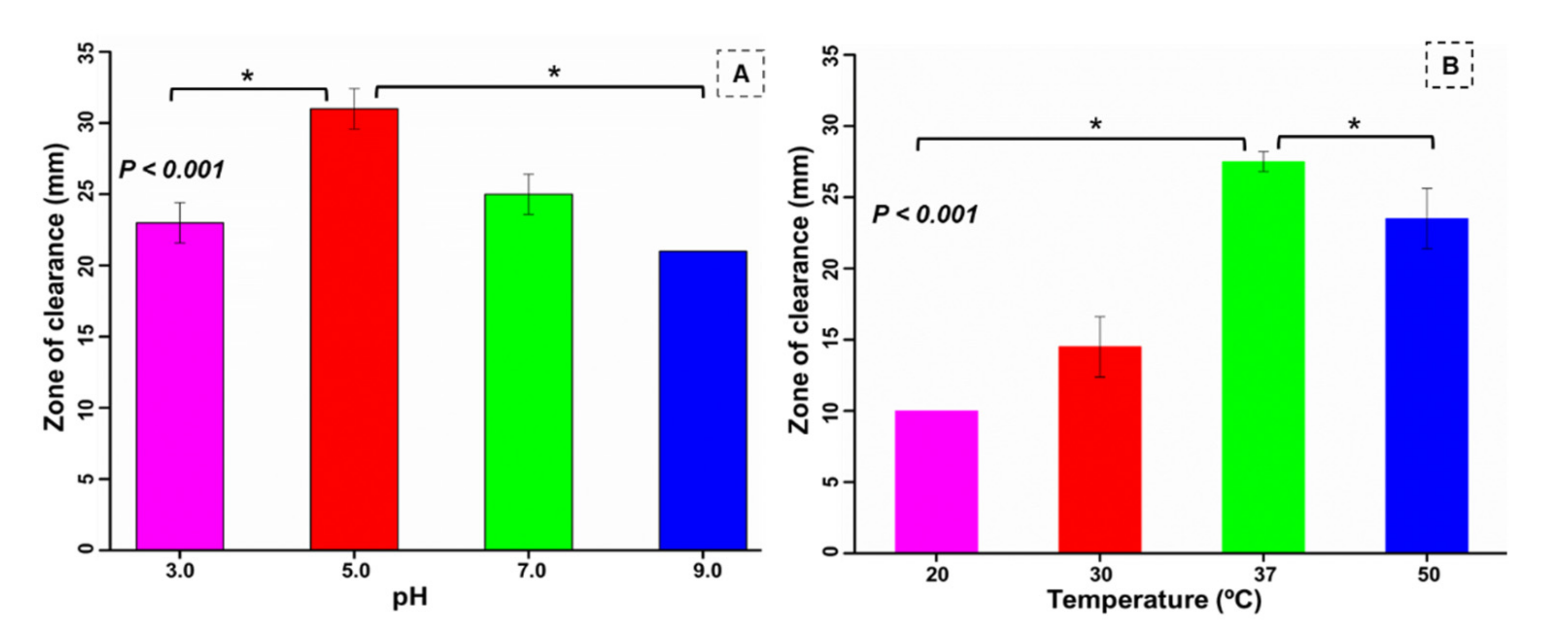
3.6. Optimization of pH and Temperature for the Bacterium
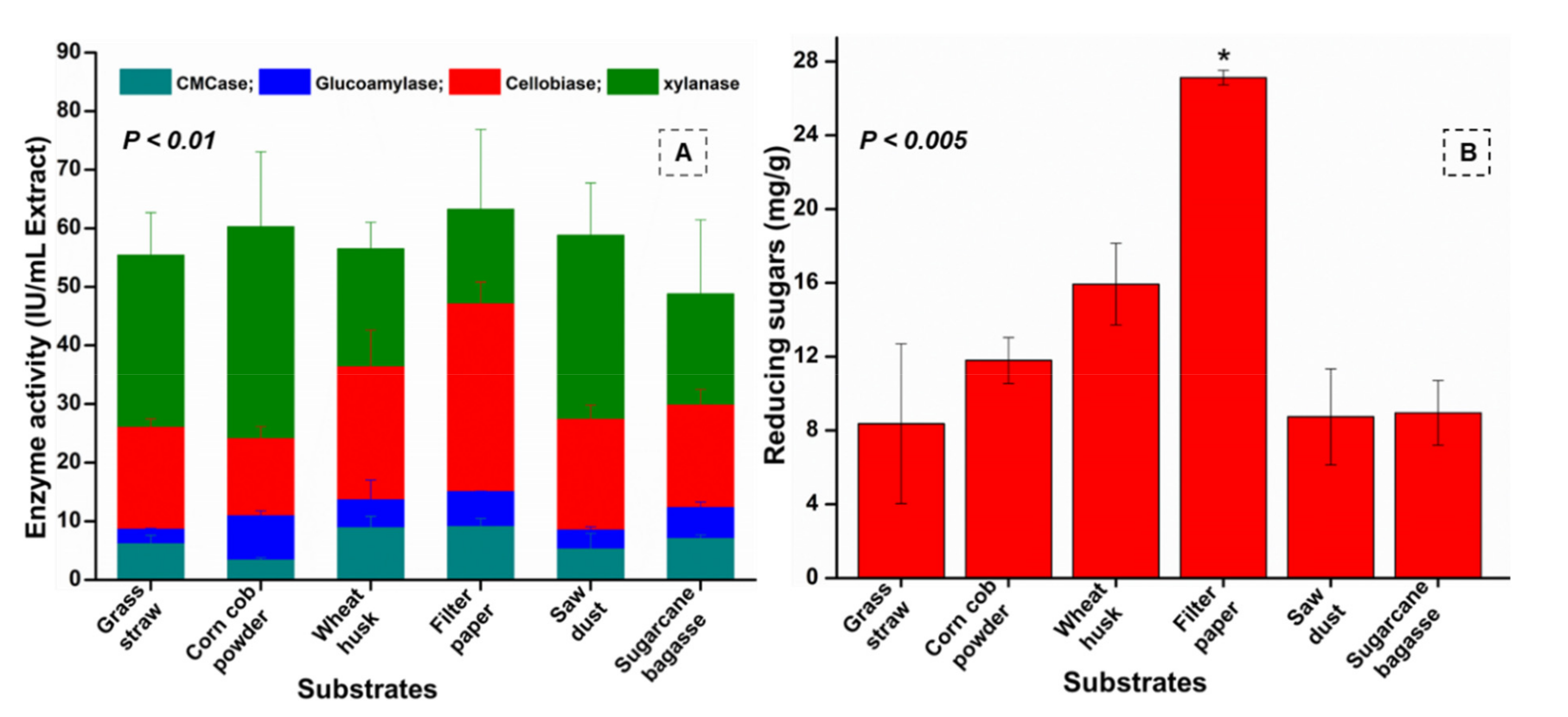
3.7. Enzyme Assays
3.8. Determination of Substrate Degradation Ratio
3.9. Scanning Electron Microscopy and Characterization of the Hydrolyzed Substrates
3.10. GS-MS Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, K.; Pei, Z.; Wang, D. Organic solvent pretreatment of lignocellulosic biomass for biofuels and biochemicals: A review. Bioresour. Technol. 2016, 199, 21–33. [Google Scholar] [CrossRef]
- Väisänen, T.; Haapala, A.; Lappalainen, R.; Tomppo, L. Utilization of agricultural and forest industry waste and residues in natural fiber-polymer composites: A review. Waste Manag. 2016, 54, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Maurya, D.P.; Singla, A.; Negi, S. An overview of key pretreatment processes for biological conversion of lignocellulosic biomass to bioethanol. 3 Biotech 2015, 5, 597–609. [Google Scholar] [CrossRef] [PubMed]
- Dashtban, M.; Schraft, H.; Qin, W. Fungal bioconversion of lignocellulosic residues; opportunities and perspectives. Int. J. Boil. 2009, 5, 578–595. [Google Scholar] [CrossRef] [PubMed]
- Maki, M.; Leung, K.T.; Qin, W. The prospects of cellulase-producing bacteria for the bioconversion of lignocellulosic biomass. Int. J. Biol. 2009, 5, 500–516. [Google Scholar] [CrossRef]
- Kalyani, D.; Lee, K.M.; Kim, T.S.; Li, J.; Dhiman, S.S.; Kang, Y.C.; Lee, J.K. Microbial consortia for saccharification of woody biomass and ethanol fermentation. Fuel 2013, 107, 815–822. [Google Scholar] [CrossRef]
- Sun, J.Z.; Scharf, M.E. Exploring and integrating cellulolytic systems of insects to advance biofuel technology. Insect Sci. 2010, 17, 163–165. [Google Scholar] [CrossRef]
- Shi, W.; Ding, S.Y.; Yuan, J.S. Comparison of insect gut cellulase and xylanase activity across different insect species with distinct food sources. Bioenergy Res. 2011, 4, 1–10. [Google Scholar] [CrossRef]
- Watanabe, H.; Tokuda, G. Animal cellulases. Cell. Mol. Life Sci. 2001, 58, 1167–1178. [Google Scholar] [CrossRef]
- Sun, J.; Ding, S.Y.; Doran-Peterson, J. Biological Conversion of Biomass for Fuels and Chemicals: Exploration from Natural Utilization Systems, 1st ed.; Royal Society of Chemistry: Cambridge, UK, 2014; pp. 1–427. [Google Scholar]
- Xie, S.X.; Syrenne, R.; Sun, S.; Yuan, J.S. Exploration of natural biomass utilization systems (NBUS) for advanced biofuel-from systems biology to synthetic design. Curr. Opin. Biotechnol. 2014, 27, 195–203. [Google Scholar] [CrossRef]
- Dar, M.A.; Shaikh, A.A.; Pawar, K.D.; Pandit, R.S. Exploring the gut of Helicoverpa armigera for cellulose degrading bacteria and evaluation of a potential strain for lignocellulosic biomass deconstruction. Proc. Biochem. 2018, 73, 142–153. [Google Scholar] [CrossRef]
- Adegboye, M.F.; Ojuederie, O.B.; Talia, P.M.; Babalol, O.O. Bioprospecting of microbial strains for biofuel production: Metabolic engineering, applications, and challenges. Biotechnol. Biofuels 2021, 14, 5. [Google Scholar] [CrossRef]
- Smith, E.H.; Whitman, R.C. Stored product pests. In Field Guide to Structural Pests; National Pest Management Association: Fairfax, VA, USA, 2007; pp. 1–764. [Google Scholar]
- Ridley, A.W.; Hereward, J.P.; Daglish, G.J.; Raghu, S.; Collins, P.J.; Walter, G.H. The spatiotemporal dynamics of Tribolium castaneum (Herbst): Adult flight and gene flow. Mol. Ecol. 2011, 20, 1635–1646. [Google Scholar] [CrossRef] [PubMed]
- Buxton, T.; Owusu, E.O.; Kim, C.S. Bioactivity of cardanol against the rust red flour beetle, Tribolium castaneum (Coleoptera: Tenebrionidae). Int. J. Trop. Insect Sci. 2018, 38, 353–361. [Google Scholar] [CrossRef]
- Padín, S.; Dal Bello, G.; Fabrizio, M. Grain loss caused by Tribolium castaneum, Sitophilus oryzae and Acanthoscelides obtectus in stored durum wheat and beans treated with Beauveria bassiana. J. Stored Prod. Res. 2002, 38, 69–74. [Google Scholar] [CrossRef]
- DeBrier, N.; Hemdane, S.; Dornez, E.; Gomand, S.V.; Delcour, J.A.; Courtin, C.M. Structure, chemical composition and enzymatic activities of pearlings and bran obtained from pearled wheat (Triticum aestivum L.) by roller milling. J. Cereal Sci. 2015, 62, 66. [Google Scholar] [CrossRef]
- Willis, J.D.; Grant, J.N.; Mazarei, M.; Kline, L.M.; Rempe, C.S.; Collins, A.G.; Turner, G.B.; Decker, S.R.; Sykes, R.W.; Davis, M.F.; et al. The TcEG1 beetle (Tribolium castaneum) cellulase produced in transgenic switchgrass is active at alkaline pH and auto-hydrolyzes biomass for increased cellobiose release. Biotechnol. Biofuels 2017, 10, 230. [Google Scholar] [CrossRef]
- Willis, J.D.; Oppert, B.; Oppert, C.; Klingeman, W.E.; Jurat-Fuentes, J.L. Identification, cloning, and expression of a GHF9 cellulase from Tribolium castaneum (Coleoptera: Tenebrionidae). J. Insect Physiol. 2011, 57, 300–306. [Google Scholar] [CrossRef]
- Shirley, D.; Oppert, C.; Reynolds, T.B.; Miracle, B.; Oppert, B.; Klingeman, W.E.; Jurat-Fuentes, J.L. Expression of an endoglucanase from Tribolium castaneum (TcEG1) in Saccharomyces cerevisiae. Insect Sci. 2014, 21, 609–618. [Google Scholar] [CrossRef]
- Rehman, F.; Aslam, M.; Tariq, M.I.; Shaheen, A.; Sami, A.J.; Naveed, N.H.; Batoo, A.I. Isolation of cellulolytic activities from Tribolium castaneum (red flour beetle). Afr. J. Biotechnol. 2009, 8, 6710–6715. [Google Scholar] [CrossRef]
- Geng, A.; Cheng, Y.; Wang, Y.; Zhu, D.; Le, Y.; Wu, J.; Xie, R.; Yuan, J.S.; Sun, J. Transcriptome analysis of the digestive system of a wood-feeding termite (Coptotermes formosanus) revealed a unique mechanism for effective biomass degradation. Biotechnol. Biofuels 2018, 11, 24. [Google Scholar] [CrossRef] [PubMed]
- Tokuda, G.; Watanabe, H.; Matsumoto, T.; Noda, H. Cellulose digestion in the wood-eating higher termite, Nasutitermes takasagoensis (shiraki): Distribution of cellulases and properties of endo-β-1,4-gIucanase. Zool. Sci. 1997, 14, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Dar, M.A.; Shaikh, A.F.; Pawar, K.D.; Xie, R.R.; Sun, J.Z.; Kanasamy, S.; Pandit, R.S. Evaluation of cellulose degrading bacteria isolated from the gut-system of cotton bollworm, Helicoverpa armigera and their potential values in biomass conversion. PeerJ 2021, 9, e11254. [Google Scholar] [CrossRef]
- Shil, R.K.; Mojumder, S.; Sadida, F.F.; Uddin, M.; Sikdar, D. Isolation and identification of cellulolytic bacteria from the gut of three phytophagus insect species. Braz. Arch. Biol. Technol. 2014, 57, 927–932. [Google Scholar] [CrossRef]
- Luo, C.; Li, Y.; Liao, H.; Yang, Y. De novo transcriptome assembly of the bamboo snout beetle, Cyrtotrachelus buqueti reveals ability to degrade lignocellulose of bamboo feedstock. Biotechnol. Biofuels 2018, 11, 292. [Google Scholar] [CrossRef]
- Lemke, T.; Ulrich, S.; Egert, M.; Friedrich, M.W.; Brune, A. Physicochemical conditions and microbial activities in the highly alkaline gut of the humus-feeding larva of Pachnoda ephippiata (Coleoptera: Scarabaeidae). Appl. Environ. Microbial. 2003, 69, 6650–6658. [Google Scholar] [CrossRef]
- Dar, M.A.; Pawar, K.D.; Pandit, R.S. Prospecting the gut fluid of giant African land snail, Achatina fulica for cellulose degrading bacteria. Int. Biodeterior. Biodegrad. 2018, 126, 103–111. [Google Scholar] [CrossRef]
- Dar, M.A.; Pawar, K.D.; Jadhav, J.P.; Pandit, R.S. Isolation of cellulolytic bacteria from the gastro-intestinal tract of Achatina fulica (Gastropoda: Pulmonata) and their evaluation for cellulose biodegradation. Int. Biodeterior. Biodegrad. 2015, 98, 73–80. [Google Scholar] [CrossRef]
- Good, N.E. The Flour Beetles of the Genus Tribolium; USDA Report; United States Department of Agriculture: Washington, DC, USA, 1936; Volume 498, pp. 1–58. [Google Scholar]
- Kirkup, D. Lucid Professional Version 2.0: Tools for identification and diagnosis. Centre for Pest Information Technology & Transfer. Ann. Bot. 2002, 89, 650–651. [Google Scholar] [CrossRef][Green Version]
- Schulthess, B.; Bloemberg, G.V.; Zbinden, R.; Böttger, E.C.; Hombach, M. Evaluation of the Bruker MALDI Biotyper for identification of gram-positive rods: Development of a diagnostic algorithm for the clinical laboratory. J. Clin. Microbiol. 2014, 52, 1089–1097. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European molecular biology open software suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Gao, Z.; Hop, V.; Yen, D.; Ando, K.; Hiyamuta, S.; Kondo, R. The production of β-glucosidases by Fusarium proliferatum NBRC109045 isolated from Vietnamese forest. AMB Express 2012, 2, 49. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farri, A.L.; Randall, R.J. Protein measurement with the folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Du, R.; Yan, J.; Li, S.; Zhang, L.; Zhang, S.; Li, J.; Zhao, G.; Qi, P. Cellulosic ethanol production by natural bacterial consortia is enhanced by Pseudoxanthomonas taiwanensis. Biotechnol. Biofuels 2015, 8, 1–10. [Google Scholar] [CrossRef]
- Dar, M.A.; Pawar, K.D.; Rajput, B.P.; Rahi, P.; Pandit, R.S. Purification of a cellulase from cellulolytic gut bacterium, Bacillus tequilensis G9 and its evaluation for valorization of agro-wastes into added value byproducts. Biocatal. Agric. Biotechnol. 2019, 20, 101219. [Google Scholar] [CrossRef]
- Galabova, D.; Tuleva, B.; Spasova, D. Permeabilization of Yerrowia lipolytica cells by Triton X-100. Enzym. Microb. Technol. 1996, 18, 18–22. [Google Scholar] [CrossRef]
- Bagewadi, Z.K.; Mulla, S.I.; Ninnekar, H.Z. Purification and characterization of endob-1,4-D-glucanase from Trichoderma harzianumstrain HZN11 and its application in production of bioethanol from sweet sorghum bagasse. 3 Biotech 2016, 6, 101. [Google Scholar] [CrossRef]
- Tsegaye, B.; Balomajumder, C.; Roy, P. Isolation and characterization of novel lignolytic, cellulolytic, and hemicellulolytic bacteria from wood-feed termite, Cyrptotermes brevis. Int. Microbiol. 2018, 22, 29–39. [Google Scholar] [CrossRef]
- Bugg, T.D.; Ahmad, M.; Hardiman, E.M.; Singh, R. The emerging role for bacteria in lignin degradation and bio-product formation. Curr. Opin. Biotechnol. 2011, 22, 394–400. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Gibbs, R.; Weinstock, G. The genome of the model beetle and pest Tribolium castaneum. Nature 2008, 452, 949–955. [Google Scholar] [CrossRef] [PubMed]
- Hou, T.S.; Chiang-Ni, C.; Teng, S.H. Current status of MALDI-TOF mass spectrometry in clinical microbiology. J. Food Drug Anal. 2019, 27, 404–414. [Google Scholar] [CrossRef] [PubMed]
- Santos, I.C.; Martin, M.S.; Carlton, D.D., Jr.; Amorim, C.L.; Castro, P.M.L.; Hildenbrand, Z.L.; Zacariah, L. MALDI-TOF/MS for the identification of cultivable organic-degrading bacteria in contaminated groundwater near unconventional natural gas extraction sites. Microorganisms 2017, 5, 47. [Google Scholar] [CrossRef] [PubMed]
- Wahl, K.L.; Wunschel, S.C.; Jarman, K.H.; Valentine, N.B.; Petersen, C.E.; Kingsley, M.T.; Zartolas, K.A.; Saenz, A.J. Analysis of microbial mixtures by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Chem. 2002, 74, 6191–6199. [Google Scholar] [CrossRef] [PubMed]
- Vasquez, A.; Forsgren, E.; Fries, I.; Paxton, R.J.; Flaberg, E.; Szekely, L.; Olofsson, T.C. Symbionts as major modulators of insect health: Lactic acid bacteria and honeybees. PLoS ONE 2012, 7, e33188. [Google Scholar] [CrossRef]
- Shao, Y.; Arias-Cordero, E.; Guo, H.; Bartram, S.; Boland, W. In vivo Pyro-SIP assessing active gut microbiota of the cotton leafworm, Spodoptera littoralis. PLoS ONE 2014, 9, e85948. [Google Scholar] [CrossRef]
- Ruiz-Rodriguez, M.; Valdivia, E.; Martin-Vivaldi, M.; Martin-Platero, A.M.; Martinez-Bueno, M.; Mendez, M.; Peralta-Sánchez, J.M.; Soler, J.J. Antimicrobial activity and genetic profile of enteroccoci isolated from hoopoes uropygial gland. PLoS ONE 2012, 7, e41843. [Google Scholar] [CrossRef]
- Pittman, G.W.; Brumbley, S.M.; Allsopp, P.G.; O’Neill, S.L. Assessment of gut bacteria for a paratransgenic approach to control Dermolepidaalbohirtum larvae. Appl. Environ. Microbiol. 2008, 74, 4036–4043. [Google Scholar] [CrossRef]
- Schloss, P.D.; Delalibera, I.; Handelsman, J.; Raffa, K.F. Bacteria associated with the guts of two wood-boring beetles: Anoplophora glabripennis and Saperda vestita (Cerambycidae). Environ. Entomol. 2006, 35, 625–629. [Google Scholar] [CrossRef]
- Dantur, K.I.; Enrique, R.; Welin, B.; Castagnaro, A.P. Isolation of cellulolytic bacteria from the intestine of Diatraea saccharalis larvae and evaluation of their capacity to degrade sugarcane biomass. AMB Express 2015, 5, 15. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Yu, J.; Wang, C.; Chen, H. Cellulolytic bacteria associated with the gut of Dendroctonusarmandilarvae (Coleoptera: Curculionidae: Scolytinae). Forests 2014, 5, 455–465. [Google Scholar] [CrossRef]
- Huang, S.; Sheng, P.; Zhang, H. Isolation and identification of cellulolytic bacteria from the gut of Holotrichia parallela larvae (Coleoptera: Scarabaeidae). Int. J. Mol. Sci. 2012, 13, 2563. [Google Scholar] [CrossRef] [PubMed]
- Pawar, K.D.; Dar, M.A.; Rajput, B.P.; Kulkarni, G.J. Enrichment and identification of cellulolytic bacteria from the gastrointestinal tract of giant African snail, Achatina fulica. Appl. Biochem. Biotechnol. 2015, 175, 1971–1980. [Google Scholar] [CrossRef] [PubMed]
- Dillon, R.J.; Dillon, V.M. The gut bacteria of insects: Nonpathogenic interactions. Annu. Rev. Entomol. 2004, 49, 71–92. [Google Scholar] [CrossRef] [PubMed]
- Suen, G.; Scott, J.J.; Aylward, F.O.; Adams, S.M.; Tringe, S.G.; Pinto-Tomas, A.A.; Foster, C.E.; Pauly, M.; Weimer, P.J.; Barry, K.W.; et al. An insect herbivore microbiome with high plant biomass-degrading capacity. PLoS Genet. 2010, 6, e1001129. [Google Scholar] [CrossRef]
- Okeke, B.C.; Lu, J. Characterization of a defined cellulolytic and xylanolytic bacterial consortium for bioprocessing of cellulose and hemicelluloses. Appl. Biochem. Biotechnol. 2011, 163, 869–881. [Google Scholar] [CrossRef]
- Ko, K.C.; Han, Y.C.; Hyun, K.J.; Geun-Joong, L.; Seung-Goo, S.J.J. A novel bifunctional endo-/exo-type cellulase from an anaerobic ruminal bacterium. Appl. Microbiol. Biotechnol. 2011, 89, 1453–1462. [Google Scholar] [CrossRef]
- Sreeja, S.J.; Jeba-Malar, P.W.; Sharmila-Joseph, F.R.; Tiburcius, S.; Immanuel, G.; Palavesam, A. Optimization of cellulase production by Bacillus altitudinis APS MSU and Bacillus licheniformis APS2 MSU, gut isolates of fish Etroplus suratensis. Int. J. Adv. Res. Technol. 2013, 2, 401–406. [Google Scholar]
- Shivaji, S.; Chaturvedi, P.; Suresh, K.; Reddy, G.S.N.; Dutt, C.B.S.; Wainwright, M.; Narlikar, J.V.; Bhargava, P.M. Bacillus aerius sp. nov., Bacillus aerophilus sp. nov., Bacillus stratosphericus sp. nov. and Bacillus altitudinis sp. nov., isolated from cryogenic tubes used for collecting air samples from high altitudes. Int. J. Syst. Evol. Microbiol. 2006, 56, 1465–1473. [Google Scholar] [CrossRef]
- Mao, S.; Lu, Z.; Zhang, C.; Lu, F.; Bie, X. Purification, characterization, and heterologous expression of a thermostable β-1,3-1,4-glucanase from Bacillus altitudinis YC-9. Appl. Biochem. Biotechnol. 2013, 169, 960–975. [Google Scholar] [CrossRef]
- Vettath, V.K.; Junqueira, A.C.M.; Uchida, A.; Purbojati, R.W.; Houghton, J.N.I.; Chénard, C.; Drautz-Moses, D.I.; Wong, A.; Kolundžija, S.; Clare, M.E.; et al. Complete genome sequence of Bacillus altitudinis type strain SGAir0031 isolated from tropical air collected in Singapore. Genome Announc. 2017, 5, e01260-17. [Google Scholar] [CrossRef]
- Sadaqat, B.; Sha, C.; Rupani, P.F.; Wang, H.; Zuo, W.; Shao, W. Man/Cel5B, a bifunctional enzyme having the highest mannanase activity in the hyperthermic environment. Front. Bioeng. Biotechnnol. 2021, 9, 637649. [Google Scholar] [CrossRef] [PubMed]
- Shakoor, S.; Saira, A.; Abdul, R. Characterization of cellulose degrading bacterium Bacillus megaterium S3, isolated from indigenous environment. Pak. J. Zool. 2013, 45, 1655–1662. [Google Scholar]
- Mathews, S.L.; Epps, M.J.; Blackburn, R.K.; Goshe, M.B.; Grunden, A.M.; Dunn, R.R. Public questions spur the discovery of new bacterial species associated with lignin bioconversion of industrial waste. R. Soc. Open Sci. 2019, 6, 180748. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Parashar, D.; Satyanarayana, T. Acidophilic microbes: Biology and applications. In Biotechnology of Extremophiles, Grand Challenges in Biology and Biotechnology; Rampelotto, P.H., Ed.; Springer: Cham, Switzerland, 2016; pp. 215–241. [Google Scholar] [CrossRef]
- Sun, X.; Liu, Z.; Qu, Y.; Li, X. The effects of wheat bran composition on the production of biomass-hydrolyzing enzymes by Penicillium decumbens. Appl. Biochem. Biotechnol. 2008, 146, 119–128. [Google Scholar] [CrossRef]
- Ma, L.; Lu, Y.; Yan, H.; Wang, X.; Yi, Y.; Shan, Y.; Liu, B.; Zhou, Y.; Lu, X. Screening of cellulolytic bacteria from rotten wood of Qinling (China) for biomass degradation and cloning of cellulases from Bacillus methylotrophicus. BMC Biotechnol. 2020, 20, 2. [Google Scholar] [CrossRef]
- Gomare, S.; Kim, H.A.; Ha, J.H.; Lee, M.W.; Park, J.M. Isolation of the polysaccharidase producing bacteria from the gut of sea snail, Batillus cornutus. Korean J. Chem. Eng. 2011, 28, 1252–1259. [Google Scholar] [CrossRef]
- Cho, M.J.; Kim, H.Y.; Shin, K.; Kim, Y.K.; Kim, Y.S.; Kim, T.J. Symbiotic adaptation of bacteria in the gut of Reticulitermes speratus: Low endo-b-1, 4-glucanase activity. Biochem. Biophys. Res. Commun. 2010, 395, 432–435. [Google Scholar] [CrossRef]
- Ferbiyanto, A.; Rusmana, I.; Raffiudin, R. Characterization and identification of cellulolytic bacteria from gut of worker Macrotermes gilvus. Hayati J. Biosci. 2015, 22, 197–200. [Google Scholar] [CrossRef]
- Islam, F.; Roy, N. Screening, purification and characterization of cellulase from cellulase producing bacteria in molasses. BMC Res. Notes 2018, 11, 445. [Google Scholar] [CrossRef] [PubMed]
- Manfredi, A.P.; Perotti, N.I.; Martínez, M.A. Cellulose degrading bacteria isolated from industrial samples and the gut of native insects from Northwest of Argentina. J. Basic Microbiol. 2015, 55, 1384–1393. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wu, S.; Wirth, S.; Hao, Y.; Wang, W.; Zou, H.; Li, W.; Wang, G. Diversity and activity of cellulolytic bacteria, isolated from the gut contents of grass carp (Ctenopharyngodon idellus) (Valenciennes) fed on Sudan grass (Sorghum sudanense) or artificial feedstuffs. Aquac. Res. 2014, 47, 153–164. [Google Scholar] [CrossRef]
- Júnior, F.L.S.; Dias, A.C.F.; Fasanella, C.C.; Taketani, R.G.; Lima, A.O.; Melo, I.S.; Andreote, F.D. Endo-and exoglucanase activities in bacteria from mangrove sediment. Braz. J. Microbiol. 2013, 44, 969–976. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Meng, F.; Peng, J.; Han, P.; Fang, F.; Ma, L.; Cao, B. Isolation and identification of a cellulolytic bacterium from the Tibetan pig’s intestine and investigation of its cellulase production. Electron. J. Biotechnol. 2014, 17, 262–267. [Google Scholar] [CrossRef]
- Singh, S.; Moholkar, V.S.; Goyal, A. Isolation, identification, and characterization of a cellulolytic Bacillus amyloliquefaciens strain SS35 from rhinoceros dung. ISRN Microbiol. 2013, 2013, 728134. [Google Scholar] [CrossRef]
- Immanuel, G.; Dhanusha, R.; Prema, P.; Palavesam, A. Effect of different growth parameters on endoglucanase enzyme activity by bacteria isolated from coir retting effluents of estuarine environment. Int. J. Environ. Sci. Technol. 2006, 3, 25–34. [Google Scholar] [CrossRef]
- Maki, M.L.; Idrees, A.; Leung, K.T.; Qin, W. Newly isolated and characterized bacteria with great application potential for decomposition of lignocellulosic biomass. J. Mol. Microbiol. Biotechnol. 2012, 22, 156–166. [Google Scholar] [CrossRef]
- Barton-Pudlik, J.; Czaja, K.; Grzymek, M.; Lipok, J. Evaluation of wood-polyethylene composites biodegradability caused by filamentous fungi. Int. Biodeterior. Biodegrad. 2017, 118, 10–18. [Google Scholar] [CrossRef]
- Xu, K.; Feng, J.; Zhong, T.; Zheng, Z.; Chen, T. Effects of volatile chemical components of wood species on mould growth susceptibility and termite attack resistance of wood plastic composites. Int. Biodeterior. Biodegrad. 2015, 100, 106–115. [Google Scholar] [CrossRef]
- Wi, S.G.; Cho, E.J.; Lee, D.S.; Lee, S.J.; Lee, Y.J.; Bae, H.J. Lignocellulose conversion for biofuel: A new pretreatment greatly improves downstream biocatalytic hydrolysis of various lignocellulosic materials. Biotechnol. Biofuels 2015, 8, 228. [Google Scholar] [CrossRef]
- Campos, A.; Marconato, J.C.; Martins-Franchetti, S.M. Biodegradation of blend films PVA/PVC, PVA/PCL in soil and soil with landfill leachate. Braz. Arch. Biol. Technol. 2011, 54, 1367–1378. [Google Scholar] [CrossRef]
- Schwarz, W.H. The cellulosome and cellulose degradation by anaerobic bacteria. Appl. Microbiol. Biotechnol. 2001, 56, 634–649. [Google Scholar] [CrossRef]
- Lu, W.J.; Wang, H.T.; Yang, S.J. Isolation and characterization of mesophilic cellulose-degrading bacteria from flower stalks-vegetable waste co-composting system. J. Gen. Appl. Microbiol. 2005, 51, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Onipe, O.O.; Afam, I.O. Jideani Beswa, D. Composition and functionality of wheat bran and its application in some cereal food products. Int. J. Food Sci. Technol. 2015, 50, 2509–2518. [Google Scholar] [CrossRef]
- Bledzki, A.K.; Mamun, A.A.; Volk, J. Physical, chemical and surface properties of wheat husk, rye husk and soft wood and their polypropylene composites. Compos. Part A Appl. Sci. Manuf. 2010, 41, 480–488. [Google Scholar] [CrossRef]
- Fatimah, S.; Sulaiman, M.R.; Hashim, O.; Arai, R.; Kosugi, T.; Abe, A.; Murata, H.Y.; Mori, Y. Characterization of parenchyma and vascular bundle of oil palm trunk as function of storage time. Lignocellulose 2012, 1, 33–44. [Google Scholar]
- Kacurakova, M.; Smith, A.C.; Gidley, M.J.; Wilson, R.H. Molecular interactions in bacterial cellulose composites studied by 1D FT-IR and dynamic 2D FT-IR spectroscopy. Carbohydr. Res. 2002, 337, 1145–1153. [Google Scholar] [CrossRef]
- Fackler, K.; Stevanic, J.S.; Ters, T.; Hinterstoisser, B.; Schwanninger, M.; Salmén, L. FTIR imaging microscopy to localize and characterize simultaneous and selective white rot decay within spruce wood cells. Holzforschung 2011, 65, 411–420. [Google Scholar] [CrossRef]
- Lionetto, F.; Sole, R.D.; Cannoletta, D.; Vasapollo, G.; Maffezzoli, A. Monitoring wood degradation during weathering by cellulose crystallinity. Materials 2012, 5, 1910. [Google Scholar] [CrossRef]
- Pandey, K.K. Study of the effect of photo-irradiation on the surface chemistry of wood. Polym. Degrad. Stab. 2005, 90, 9–20. [Google Scholar] [CrossRef]
- Wang, L.; Han, G. Yuanming Zhang Comparative study of composition, structure and properties of Apocynum venetum fibers under different pretreatments. Carbohydr. Polym. 2007, 69, 391–397. [Google Scholar] [CrossRef]
- Colom, X.; Carrillo, F. Crystallinity changes in lyocell and viscose-type fibers by caustic treatment. Eur. Polym. J. 2002, 38, 2225–2230. [Google Scholar] [CrossRef]
- Binod, P.; Satyanagalakshmi, K.; Sindhu, R.; Janu, K.U.; Sukumaran, R.K.; Pandey, A. Short duration microwave assisted pretreatment enhances the enzymatic saccharification and fermentable sugar yield from sugarcane bagasse. Renew. Energy 2012, 37, 109–116. [Google Scholar] [CrossRef]
- Kar, K.; Cheng, W. Using Mass Spectrometry to Detect Ethanol and Acetaldehyde Emissions from a Direct Injection Spark Ignition Engine Operating on Ethanol/Gasoline Blends; 2011–1–11959; SAE International in United States: Warrendale, PA, USA, 2011; pp. 1–14. [Google Scholar] [CrossRef]
- Sun, Z.; Liu, K.; Zhang, J.; Zhang, Y.; Xu, K.; Yu, D.; Wang, J.; Hu, L.; Chen, L.; Li, C. IAA producing Bacillus altitudinis alleviates iron stress in Triticum aestivum L. seedling by both bioleaching of iron and up-regulation of genes encoding ferritins. Plant Soil 2017, 419, 1–11. [Google Scholar] [CrossRef]
- Harish, K.; Reddy, Y.; Srijana, M.; Reddy, D.M.; Reddy, G. Coculture fermentation of banana agro-waste to ethanol by cellulolytic thermophilic Clostridium thermocellum CT2. Afr. J. Biotechnol. 2010, 9, 1926–1934. [Google Scholar] [CrossRef]
- Qian, M.; Tian, S.; Li, X.; Zhang, J.; Pan, Y.; Yang, X. Ethanol production from dilute-acid softwood hydrolysate by co-culture. Appl. Biochem. Biotechnol. 2006, 134, 273–283. [Google Scholar] [CrossRef]

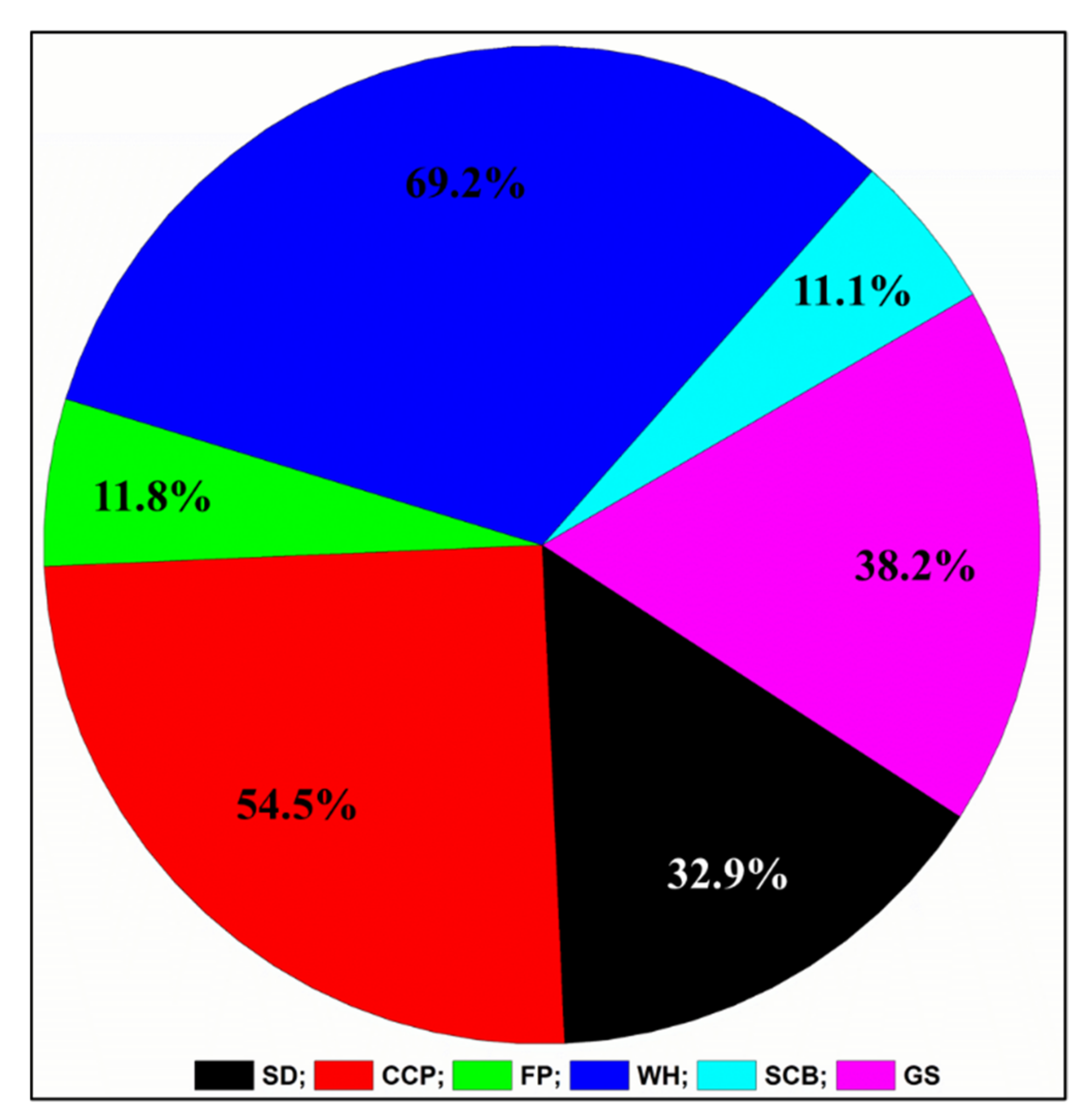
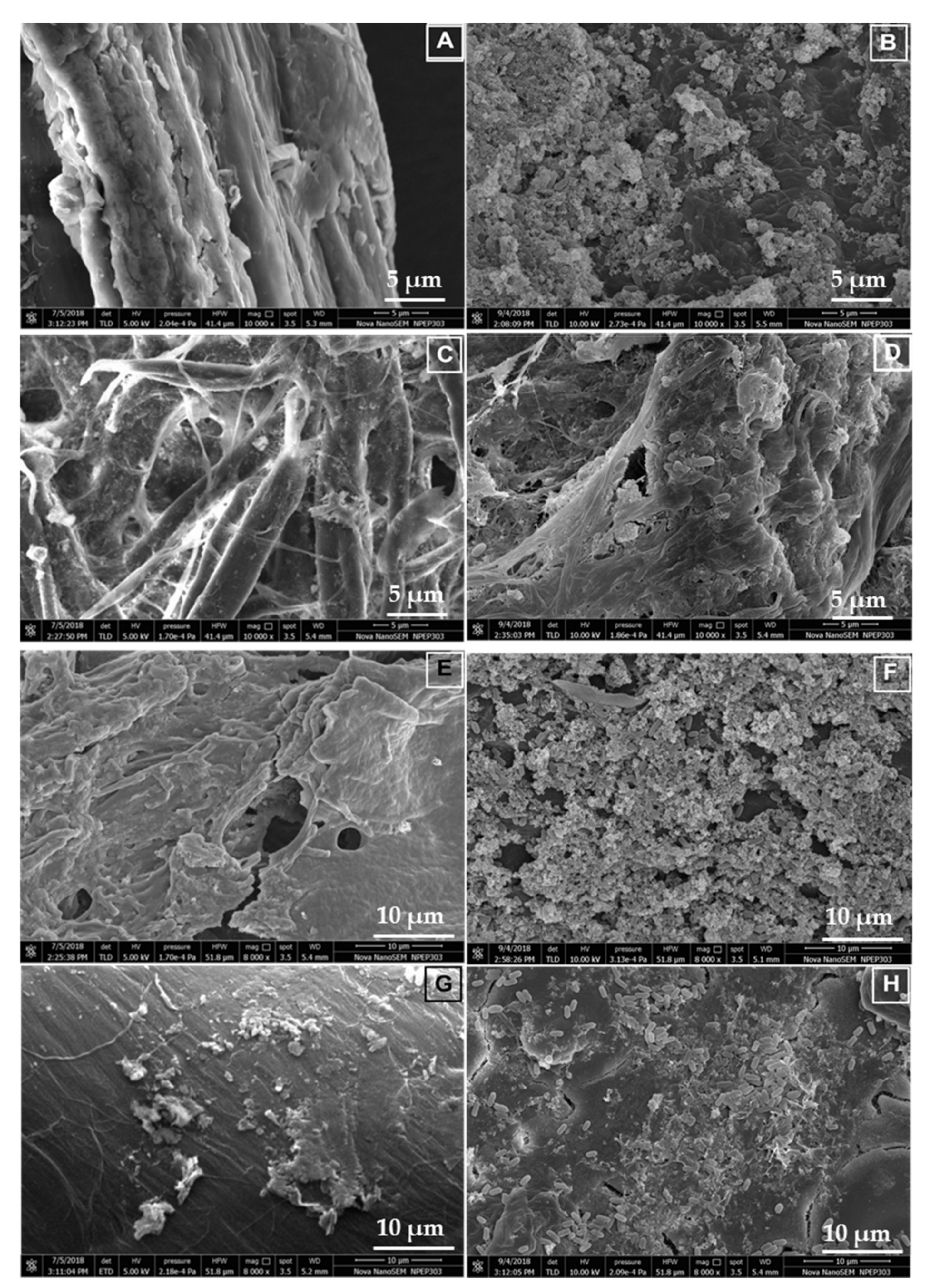
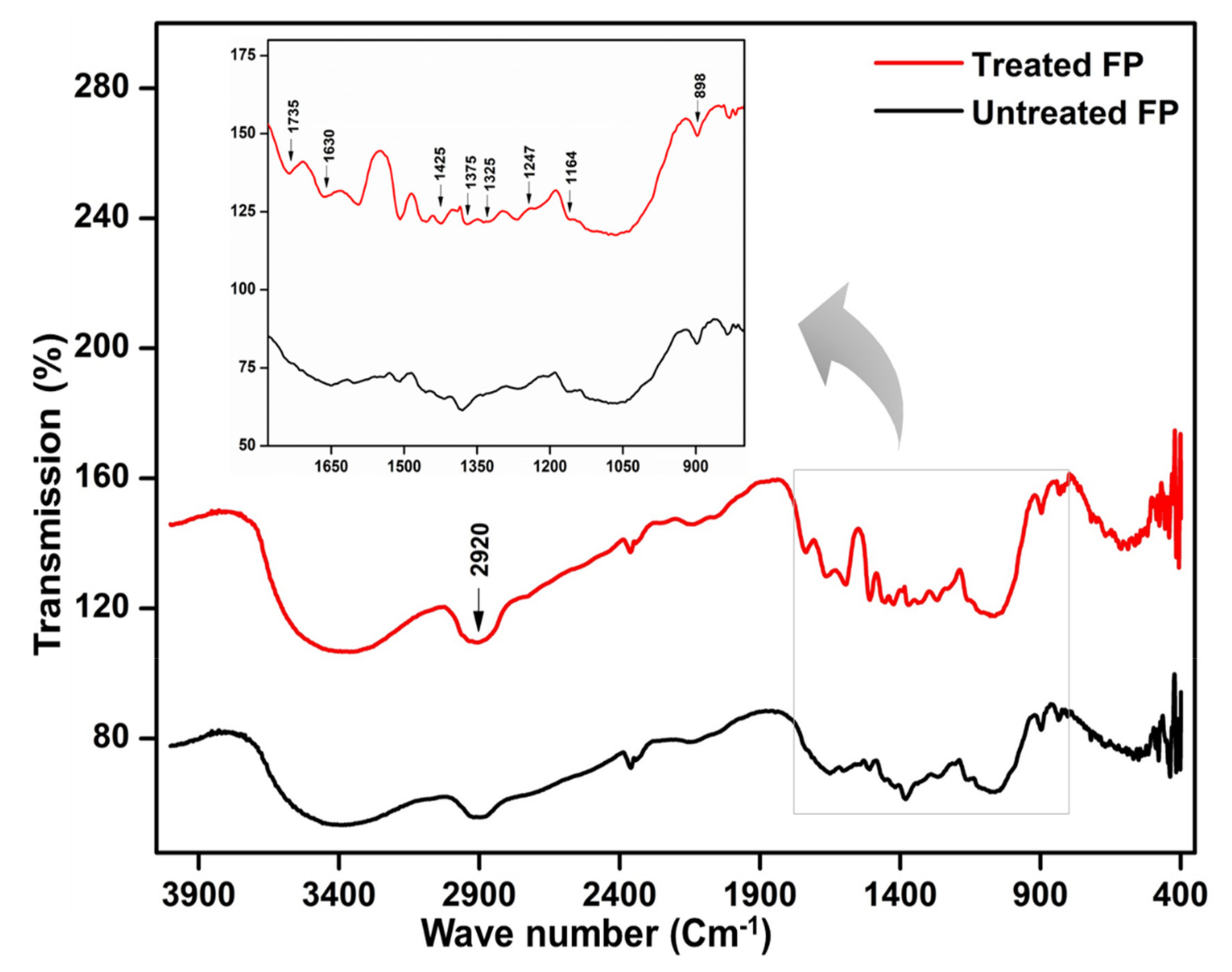
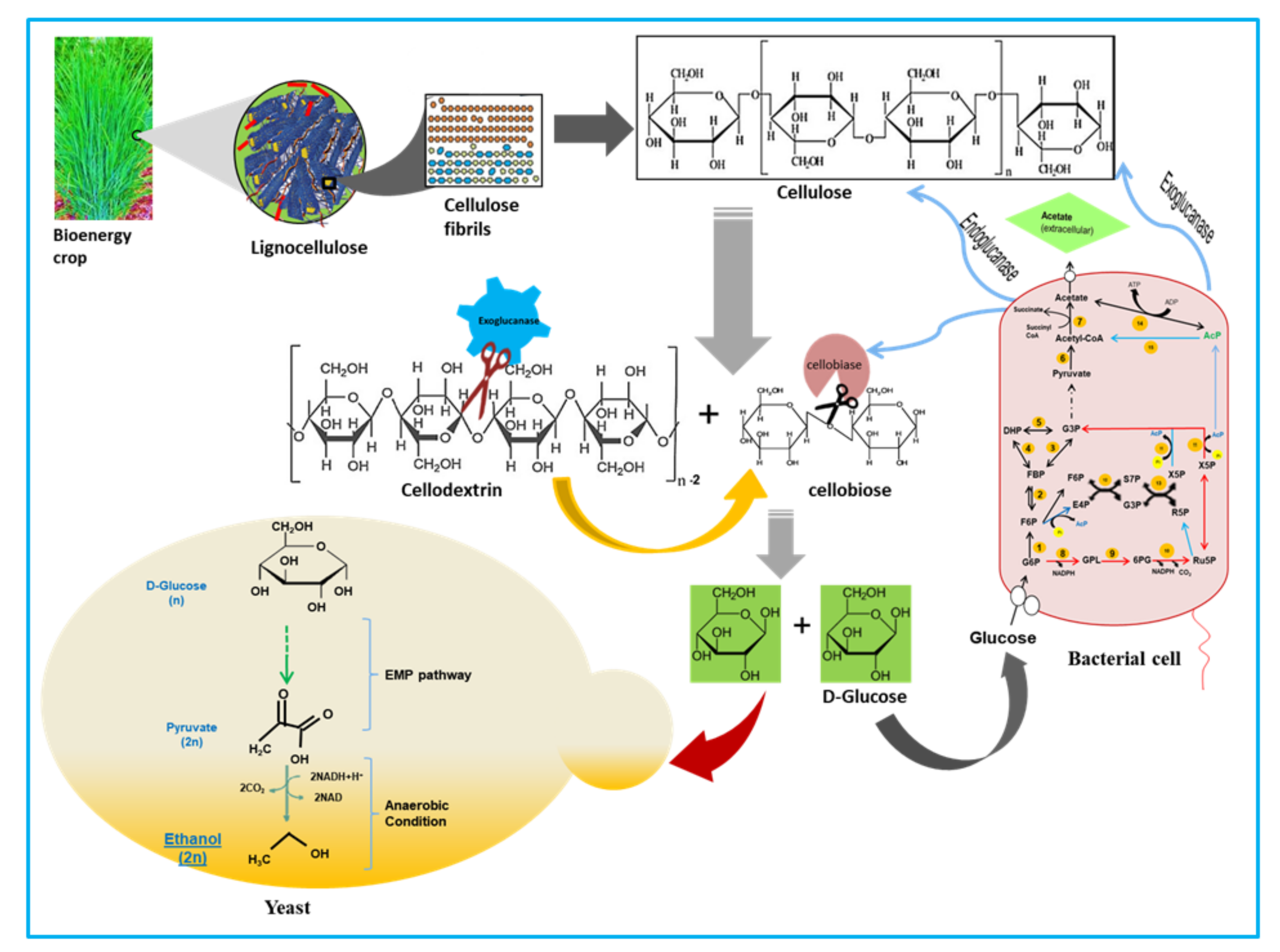
| Sr. No. | Isolate Code | Best Hit | MALDI-TOF Score |
|---|---|---|---|
| 1 | TC5 | Achromobacter spanius | 2.057 |
| 2 | TC7 | Escherichia hermannii | 1.824 |
| 3 | TC10 | Bacillus subtilis | 1.946 |
| 4 | TC11 | Bacillus sp. | 1.806 |
| 5 | TC16 | Enterococcus faecalis | 1.855 |
| 6 | TC18 | Citrobacter freundii | 1.851 |
| 7 | TC24 | Enterococcus faecalis | 2.157 |
| 8 | TC27 | Achromobacter insolitus | 2.244 |
| 9 | TC39 | Bacillus subtilis | 1.802 |
| 10 | TC41 | Kluyvera georgiana | 2.244 |
| 11 | TC55 | Bacillus sp. | 2.218 |
| 12 | TC67 | Cronobacter sakazakii | 1.828 |
| 13 | TC70 | Escherichia coli | 1.883 |
| 14 | RSP75 | Bacillus sp. | 2.207 |
| 15 | TC91 | Kluyvera ascorbata | 1.915 |
| Peak No. | Compound Name | Area (%) | Retention Index | Rt (min) |
|---|---|---|---|---|
| 1 | Ethanol | 0.78 | 45 | 2.523 |
| 2 | Carbon dioxide | 7.39 | 44 | 1.490 |
| 3 | Acetic acid | 18.31 | 576 | 2.025 |
| 4 | Acetone alcohol | 14.57 | 698 | 2.275 |
| 5 | Acetyldehyde | 6.00 | 29 | 3.115 |
| 6 | 2-Propanone | 1.09 | 698 | 2.472 |
| Bacteria | NCBI Accession | Substrate Clearance Zone (mm) | Cellulase Activity IU/mL (or mg) Extract | Xylanase Activity IU/mL Extract | Reference |
|---|---|---|---|---|---|
| B. altitudinis RSP75 | MW559543 | 28 | 47.1 ± 3 | 60.2 ± 2 | This study |
| B. subtilis 1AJ3 | MG062801 | 19 | 0.04 | ND | [72] |
| B. methylotrophicus 1EJ7 | MG062824 | 19 | 0.025 | ND | |
| Bacillus sp. JMP-A | HM776393 | 11 | 27.10 | ND | [73] |
| Bacillus sp. NT4 | GU458276 | ND | ~2.5 | ND | [74] |
| Bacillus megaterium RU4 | NA | 14.5 | ND | ND | [75] |
| Paenibacillus sp. C1 | NA | NA | 0.9 | ND | [76] |
| Bacillus anthracis AR426 | LN829572 | ND | 0.36 ± 0.04 | ND | [77] |
| Cohnella formosensis AR92 | FJ976043 | ND | 0.15 ± 0.03 | ND | |
| B. tequilensis G9 | KR866144 | 25 | 598.07 | 74.02 | [29] |
| Bacillus megaterium SCMC89 | KF358455 | >8 | ND | ND | [78] |
| Exiguobacterium marinum TN25 | NA | 16 | ND | ND | [79] |
| B. subtilis BY-2 | KC414931 | NA | 1.52 | ND | [80] |
| Bacillus amyloliquefaciens SS35 | AB679994 | NA | 0.079 | ND | [81] |
| Cellulomonas sp. | NA | NA | 0.0336 | ND | [82] |
| Bacillus sp. | NA | NA | 0.0196 | ND | |
| Micrococcus sp. | NA | NA | 0.0152 | ND |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dar, M.A.; Dhole, N.P.; Xie, R.; Pawar, K.D.; Ullah, K.; Rahi, P.; Pandit, R.S.; Sun, J. Valorization Potential of a Novel Bacterial Strain, Bacillus altitudinis RSP75, towards Lignocellulose Bioconversion: An Assessment of Symbiotic Bacteria from the Stored Grain Pest, Tribolium castaneum. Microorganisms 2021, 9, 1952. https://doi.org/10.3390/microorganisms9091952
Dar MA, Dhole NP, Xie R, Pawar KD, Ullah K, Rahi P, Pandit RS, Sun J. Valorization Potential of a Novel Bacterial Strain, Bacillus altitudinis RSP75, towards Lignocellulose Bioconversion: An Assessment of Symbiotic Bacteria from the Stored Grain Pest, Tribolium castaneum. Microorganisms. 2021; 9(9):1952. https://doi.org/10.3390/microorganisms9091952
Chicago/Turabian StyleDar, Mudasir A., Neeraja P. Dhole, Rongrong Xie, Kiran D. Pawar, Kalim Ullah, Praveen Rahi, Radhakrishna S. Pandit, and Jianzhong Sun. 2021. "Valorization Potential of a Novel Bacterial Strain, Bacillus altitudinis RSP75, towards Lignocellulose Bioconversion: An Assessment of Symbiotic Bacteria from the Stored Grain Pest, Tribolium castaneum" Microorganisms 9, no. 9: 1952. https://doi.org/10.3390/microorganisms9091952
APA StyleDar, M. A., Dhole, N. P., Xie, R., Pawar, K. D., Ullah, K., Rahi, P., Pandit, R. S., & Sun, J. (2021). Valorization Potential of a Novel Bacterial Strain, Bacillus altitudinis RSP75, towards Lignocellulose Bioconversion: An Assessment of Symbiotic Bacteria from the Stored Grain Pest, Tribolium castaneum. Microorganisms, 9(9), 1952. https://doi.org/10.3390/microorganisms9091952







