Integrated Analysis of miRNA-mRNA Network Reveals Different Regulatory Patterns in the Endometrium of Meishan and Duroc Sows during Mid-Late Gestation
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Sample Collection and RNA Extraction
2.2. Library Preparation for mRNA Sequencing and Data Analysis
2.3. Library Preparation for MicroRNA Sequencing and Data Analysis
2.4. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Enrichment Analyses
2.5. Integrated miRNA-mRNA Analysis
2.6. Quantitative Polymerase Chain Reaction
2.7. Cell Culture and Transfection
2.8. Dual Luciferase Assay
2.9. Statistical Analysis
3. Results
3.1. Overview of Sequencing Data
3.2. Identifying Differentially Expressed mRNAs and miRNAs
3.3. Functional Analysis of DEGs and DE miRNA Target Genes
3.4. miRNA-mRNA Interaction Analysis
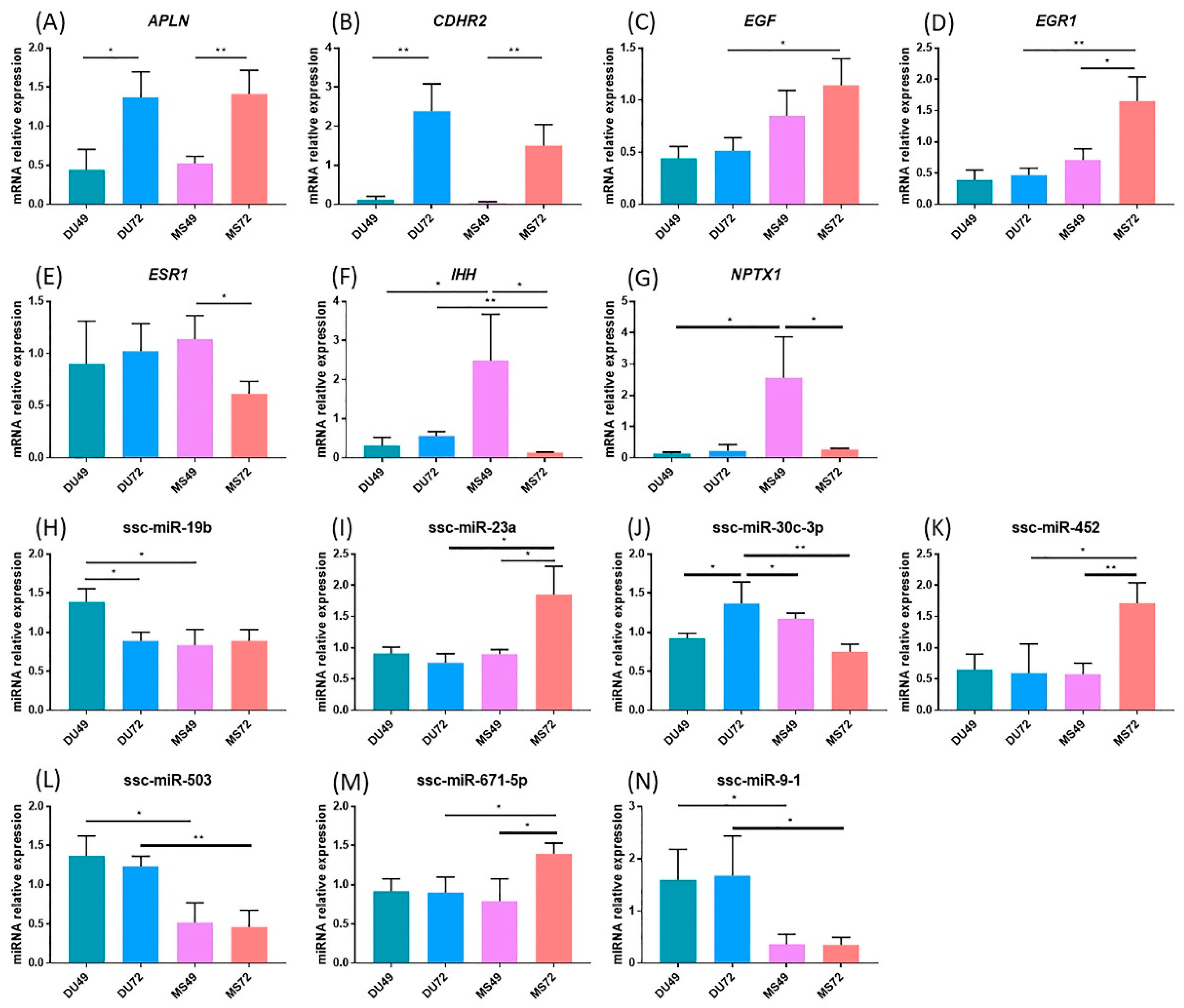
3.5. qPCR Validation for DEGs and DE miRNAs
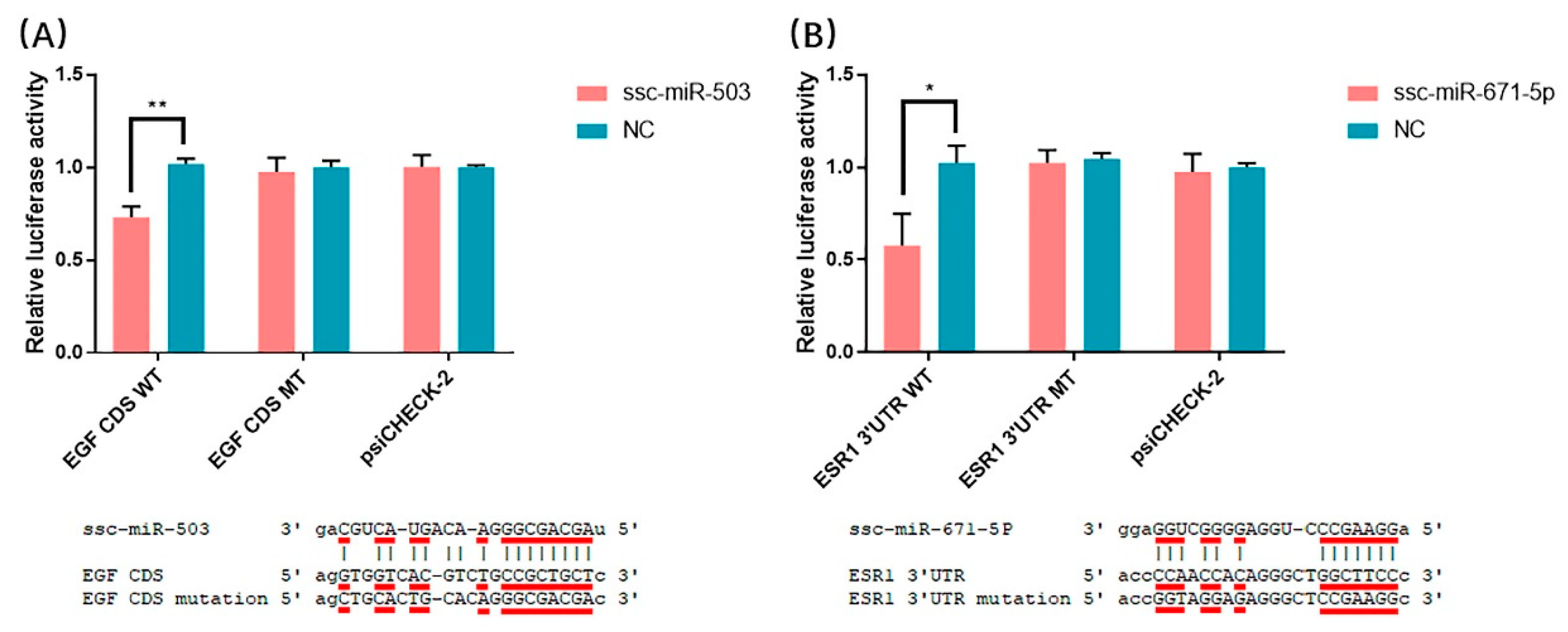
3.6. Validation of miRNA-mRNA Interactions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kridli, R.T.; Khalaj, K.; Bidarimath, M.; Tayade, C. Placentation, maternal-fetal interface, and conceptus loss in swine. Theriogenology 2016, 85, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Freking, B.A.; Lents, C.A.; Vallet, J.L. Selection for uterine capacity improves lifetime productivity of sows. Anim. Reprod. Sci. 2016, 167, 16–21. [Google Scholar] [CrossRef]
- Haley, C.S.; Lee, G.J.; Ritchie, M. Comparative reproductive performance in Meishan and Large White pigs and their crosses. Anim. Sci. 1995, 60, 259–267. [Google Scholar] [CrossRef]
- Youngs, C.R.; Ford, S.P.; McGinnis, L.K.; Anderson, L.H. Investigations into the control of litter size in swine: I. Comparative studies on in vitro development of Meishan and Yorkshire preimplantation embryos. J. Anim. Sci. 1993, 71, 1561–1565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, L.H.; Christenson, L.K.; Christenson, R.K.; Ford, S.P. Investigations into the control of litter size in swine: II. Comparisons of morphological and functional embryonic diversity between Chinese and American breeds. J. Anim. Sci. 1993, 71, 1566–1571. [Google Scholar] [CrossRef] [PubMed]
- Biensen, N.J.; Wilson, M.E.; Ford, S.P. The impact of either a Meishan or Yorkshire uterus on Meishan or Yorkshire fetal and placental development to days 70, 90, and 110 of gestation. J. Anim. Sci. 1998, 76, 2169–2176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nina, J.; Wilson, M.E.; Ford, S. The impacts of uterine environment and fetal genotype on conceptus size and placental vascularity during late gestation in pigs. J. Anim. Sci. 1999, 77, 954–959. [Google Scholar] [CrossRef]
- Bidarimath, M.; Tayade, C. Pregnancy and spontaneous fetal loss: A pig perspective. Mol. Reprod. Dev. 2017, 84, 856–869. [Google Scholar] [CrossRef] [Green Version]
- Ford, S. Embryonic and fetal development in different genotypes in pigs. J. Reprod. Fertil. Suppl. 1997, 52, 165–176. [Google Scholar]
- Leymaster, K.; Vallet, J.; Christenson, R. Number of fetuses and conceptus growth throughout gestation in lines of pigs selected for ovulation rate or uterine capacity. J. Anim. Sci. 2007, 85, 2093–2103. [Google Scholar] [CrossRef] [Green Version]
- Waclawik, A.; Kaczmarek, M.M.; Blitek, A.; Kaczynski, P.; Ziecik, A.J. Embryo-maternal dialogue during pregnancy establishment and implantation in the pig. Mol. Reprod. Dev. 2017, 84, 842–855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christenson, R.K.; Vallet, J.L.; Leymaster, K.; Young, L.D. Uterine function in Meishan pigs. J. Reprod. Fertil. Suppl. 1993, 48, 279–289. [Google Scholar] [PubMed]
- Brennecke, J.; Hipfner, D.R.; Stark, A.; Russell, R.B.; Cohen, S.M. Bantam Encodes a Developmentally Regulated microRNA that Controls Cell Proliferation and Regulates the Proapoptotic Gene hid in Drosophila. Cell 2003, 113, 25–36. [Google Scholar] [CrossRef] [Green Version]
- Dong, H.; Lei, J.; Ding, L.; Wen, Y.; Ju, H.; Zhang, X. MicroRNA: Function, detection, and bioanalysis. Chem. Rev. 2013, 113, 6207–6233. [Google Scholar] [CrossRef] [PubMed]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Krawczynski, K.; Najmula, J.; Bauersachs, S.; Kaczmarek, M.M. MicroRNAome of porcine conceptuses and trophoblasts: Expression profile of micrornas and their potential to regulate genes crucial for establishment of pregnancy. Biol. Reprod. 2015, 92, 21. [Google Scholar] [CrossRef] [PubMed]
- Su, L.; Liu, R.; Cheng, W.; Zhu, M.; Li, X.; Zhao, S.; Yu, M. Expression patterns of microRNAs in porcine endometrium and their potential roles in embryo implantation and placentation. PLoS ONE 2014, 9, e87867. [Google Scholar] [CrossRef]
- Liu, R.; Wang, M.; Su, L.; Li, X.; Zhao, S.; Yu, M. The Expression Pattern of MicroRNAs and the Associated Pathways Involved in the Development of Porcine Placental Folds That Contribute to the Expansion of the Exchange Surface Area. Biol. Reprod. 2015, 93, 62. [Google Scholar] [CrossRef]
- Wang, K.; Yang, K.; Xu, Q.; Liu, Y.; Li, W.; Bai, Y.; Wang, J.; Ding, C.; Liu, X.; Tang, Q.; et al. Protein expression profiles in Meishan and Duroc sows during mid-gestation reveal differences affecting uterine capacity, endometrial receptivity, and the maternal-fetal Interface. BMC Genom. 2019, 20, 991. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, S.; Liu, M.; Zhang, A.; Wu, Z.; Zhang, Z.; Li, J. Differential gene expression in the endometrium on gestation day 12 provides insight into sow prolificacy. BMC Genom. 2013, 14, 45. [Google Scholar] [CrossRef] [Green Version]
- Gu, T.; Zhu, M.-J.; Schroyen, M.; Qu, L.; Nettleton, D.; Kuhar, D.; Lunney, J.K.; Ross, J.W.; Zhao, S.-H.; Tuggle, C.K. Endometrial gene expression profiling in pregnant Meishan and Yorkshire pigs on day 12 of gestation. BMC Genom. 2014, 15, 156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.; Liu, R.; Su, L.; Xiao, Q.; Yu, M. Transcriptome Analysis Revealed the Embryo-Induced Gene Expression Patterns in the Endometrium from Meishan and Yorkshire Pigs. Int. J. Mol. Sci. 2015, 16, 22692–22710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, H.; Wang, H.; Wang, Y.; Liu, C.; Wang, C.; Guo, J. Transcriptomic Analysis of the Porcine Endometrium during Embryo Implantation. Genes 2015, 6, 1330–1346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, Y.; Smyth, G.K.; Shi, W. Featurecounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [Green Version]
- Friedlander, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Enright, A.J.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D.S. MicroRNA targets in Drosophila. Genome Biol. 2003, 5, R1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Onteru, S.K.; Fan, B.; Du, Z.Q.; Garrick, D.J.; Stalder, K.J.; Rothschild, M.F. A whole-genome association study for pig reproductive traits. Anim. Genet. 2012, 43, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, L.; Li, Y.; Li, W.; Luo, W.; Cheng, D.; Yan, H.; Ma, X.; Liu, X.; Song, X.; et al. Data mining in networks of differentially expressed genes during sow pregnancy. Int. J. Biol. Sci. 2012, 8, 548–560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, W.; Bae, H.; Bazer, F.W.; Song, G. Stimulatory effects of fibroblast growth factor 2 on proliferation and migration of uterine luminal epithelial cells during early pregnancy. Biol. Reprod. 2016, 96, 185–198. [Google Scholar] [CrossRef] [Green Version]
- Hiramoto, K.; Yamate, Y.; Sato, E.F. Gp91phox NADPH oxidase modulates litter size by regulating mucin1 in the uterus of mice. Syst. Biol. Reprod. Med. 2017, 63, 130–139. [Google Scholar] [CrossRef] [Green Version]
- Cordoba, S.; Balcells, I.; Castello, A.; Ovilo, C.; Noguera, J.L.; Timoneda, O.; Sanchez, A. Endometrial gene expression profile of pregnant sows with extreme phenotypes for reproductive efficiency. Sci. Rep. 2015, 5, 14416. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Ye, J.; Han, X.; Qiao, R.; Li, X.; Lv, G.; Wang, K. Whole-genome sequencing identifies potential candidate genes for reproductive traits in pigs. Genomics 2020, 112, 199–206. [Google Scholar] [CrossRef]
- Chen, Z.; Ye, S.; Teng, J.; Diao, S.; Yuan, X.; Chen, Z.; Zhang, H.; Li, J.; Zhang, Z. Genome-wide association studies for the number of animals born alive and dead in duroc pigs. Theriogenology 2019, 139, 36–42. [Google Scholar] [CrossRef]
- Wang, Y.; Ding, X.; Tan, Z.; Xing, K.; Yang, T.; Wang, Y.; Sun, D.; Wang, C. Genome-wide association study for reproductive traits in a Large White pig population. Anim. Genet. 2018, 49, 127–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.Q.; Li, F.E.; Deng, C.Y. Short Communication: Molecular cloning and expression pattern of the porcine 5-aminolevulinate synthase 1 (ALAS1) gene and its association with reproductive traits. Genet. Mol. Res. 2016, 15. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Wang, K.; Yang, Q.; Zhou, J.; Chen, D.; Ma, J.; Tang, Q.; Jin, L.; Xiao, W.; Jiang, A.; et al. Identifying SNPs and candidate genes for three litter traits using single-step GWAS across six parities in Landrace and Large White pigs. Physiol. Genom. 2018, 50, 1026–1035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Fu, J.; Wang, A. Expression of genes involved in progesterone receptor paracrine signaling and their effect on litter size in pigs. J. Anim. Sci. Biotechnol. 2016, 7, 31. [Google Scholar] [CrossRef] [Green Version]
- Henkes, L.E.; Pru, J.K.; Ashley, R.L.; Anthony, R.V.; Veeramachaneni, D.N.; Gates, K.C.; Hansen, T.R. Embryo mortality in Isg15-/- mice is exacerbated by environmental stress. Biol. Reprod. 2015, 92, 36. [Google Scholar] [CrossRef] [Green Version]
- Mucha, A.; Ropka-Molik, K.; Piorkowska, K.; Tyra, M.; Oczkowicz, M. Effect of EGF, AREG and LIF genes polymorphisms on reproductive traits in pigs. Anim. Reprod. Sci. 2013, 137, 88–92. [Google Scholar] [CrossRef]
- Sharma, S.; Sharma, P.M.; Mistry, D.S.; Chang, R.J.; Olefsky, J.M.; Mellon, P.L.; Webster, N.J. PPARG regulates gonadotropin-releasing hormone signaling in LbetaT2 cells in vitro and pituitary gonadotroph function in vivo in mice. Biol. Reprod. 2011, 84, 466–475. [Google Scholar] [CrossRef]
- Metodiev, S.; Thekkoot, D.M.; Young, J.M.; Onteru, S.; Rothschild, M.F.; Dekkers, J.C.M. A whole-genome association study for litter size and litter weight traits in pigs. Livest. Sci. 2018, 211, 87–97. [Google Scholar] [CrossRef]
- Rothschild, M.; Jacobson, C.; Vaske, D.; Tuggle, C.; Wang, L.; Short, T.; Eckardt, G.; Sasaki, S.; Vincent, A.; McLaren, D.; et al. The estrogen receptor locus is associated with a major gene influencing litter size in pigs. Proc. Natl. Acad. Sci. USA 1996, 93, 201. [Google Scholar] [CrossRef] [Green Version]
- Ashworth, C.J.; Haley, C.S.; Aitken, R.P.; Wilmut, I. Embryo survival and conceptus growth after reciprocal embryo transfer between Chinese Meishan and Landrace x Large White gilts. J. Reprod. Fertil. 1990, 90, 595–603. [Google Scholar] [CrossRef] [Green Version]
- Wessels, J.M.; Edwards, A.K.; Khalaj, K.; Kridli, R.T.; Bidarimath, M.; Tayade, C. The microRNAome of pregnancy: Deciphering miRNA networks at the maternal-fetal interface. PLoS ONE 2013, 8, e72264. [Google Scholar] [CrossRef] [PubMed]
- Rah, H.; Chung, K.W.; Ko, K.H.; Kim, E.S.; Kim, J.O.; Sakong, J.H.; Kim, J.H.; Lee, W.S.; Kim, N.K. miR-27a and miR-449b polymorphisms associated with a risk of idiopathic recurrent pregnancy loss. PLoS ONE 2017, 12, e0177160. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Han, X.; Huang, Z.; Yu, M.; Zhang, Y.; Fan, Y.; Xu, B.; Zhou, K.; Song, L.; Wang, X.; et al. Maternal pentachlorophenol exposure induces developmental toxicity mediated by autophagy on pregnancy mice. Ecotoxicol. Environ. Saf. 2019, 169, 829–836. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Zhang, X.; Cai, Z.; Zhou, J.; Cao, R.; Zhao, Y.; Chen, Z.; Wang, D.; Ruan, W.; Zhao, Q.; et al. A novel class of microRNA-recognition elements that function only within open reading frames. Nat. Struct. Mol. Biol. 2018, 25, 1019–1027. [Google Scholar] [CrossRef] [PubMed]
- Dunn, I.F.; Heese, O.; Black, P.M. Growth Factors in Glioma Angiogenesis: FGFs, PDGF, EGF, and TGFs. J. Neuro-Oncol. 2000, 50, 121–137. [Google Scholar] [CrossRef]
- Zhang, L.; Takara, K.; Yamakawa, D.; Kidoya, H.; Takakura, N. Apelin as a marker for monitoring the tumor vessel normalization window during antiangiogenic therapy. Cancer Sci. 2016, 107, 36–44. [Google Scholar] [CrossRef] [Green Version]
- Van Mieghem, T.; Doherty, A.; Baczyk, D.; Drewlo, S.; Baud, D.; Carvalho, J.; Kingdom, J. Apelin in Normal Pregnancy and Pregnancies Complicated by Placental Insufficiency. Reprod. Sci. 2016, 23, 1037–1043. [Google Scholar] [CrossRef]
- Wen, Y.; Chen, R.; Zhu, C.; Qiao, H.; Liu, Y.; Ji, H.; Miao, J.; Chen, L.; Liu, X.; Yang, Y. MiR-503 suppresses hypoxia-induced proliferation, migration and angiogenesis of endothelial progenitor cells by targeting Apelin. Peptides 2018, 105, 58–65. [Google Scholar] [CrossRef]
- Yang, W.; Lu, Z.; Zhi, Z.; Liu, L.; Deng, L.; Jiang, X.; Pang, L. Increased miRNA-518b inhibits trophoblast migration and angiogenesis by targeting EGR1 in early embryonic arrest. Biol. Reprod. 2019, 101, 664–674. [Google Scholar] [CrossRef]
- Rocha, S.F.; Schiller, M.; Jing, D.; Li, H.; Butz, S.; Vestweber, D.; Biljes, D.; Drexler, H.C.; Nieminen-Kelha, M.; Vajkoczy, P.; et al. Esm1 modulates endothelial tip cell behavior and vascular permeability by enhancing VEGF bioavailability. Circ. Res. 2014, 115, 581–590. [Google Scholar] [CrossRef] [Green Version]
- De Jong, O.G.; van der Waals, L.M.; Kools, F.R.W.; Verhaar, M.C.; van Balkom, B.W.M. Lysyl oxidase-like 2 is a regulator of angiogenesis through modulation of endothelial-to-mesenchymal transition. J. Cell Physiol. 2019, 234, 10260–10269. [Google Scholar] [CrossRef] [Green Version]
- Geisert, R.D.; Lucy, M.C.; Whyte, J.J.; Ross, J.W.; Mathew, D.J. Cytokines from the pig conceptus: Roles in conceptus development in pigs. J. Anim. Sci. Biotechnol. 2014, 5, 51. [Google Scholar] [CrossRef] [Green Version]
- Mathew, D.J.; Lucy, M.C.; Geisert, R.D. Interleukins, interferons, and establishment of pregnancy in pigs. Reproduction 2016, 151, R111–R122. [Google Scholar] [CrossRef] [Green Version]
- Schanz, A.; Baston-Büst, D.M.; Heiss, C.; Beyer, I.M.; Krüssel, J.S.; Hess, A.P. Interferon stimulated gene 15 expression at the human embryo−maternal interface. Arch. Gynecol. Obstet. 2014, 290, 783–789. [Google Scholar] [CrossRef]
- Zhao, S.; Wu, Y.; Gao, H.; Evans, A.; Zeng, S.-M. Roles of interferon-stimulated gene 15 protein in bovine embryo development. Reprod. Fertil. Dev. 2017, 29, 1209–1216. [Google Scholar] [CrossRef]
- Wilson, M.E.; Ford, S.P. Differences in trophectoderm mitotic rate and P450 17alpha-hydroxylase expression between late preimplantation Meishan and Yorkshire conceptuses. Biol. Reprod. 1997, 56, 380–385. [Google Scholar] [CrossRef] [Green Version]
- Wilson, M.E.; Ford, S.P. Effect of estradiol-17β administration during the time of conceptus elongation on placental size at term in Meishan pigs. J. Anim. Sci. 2000, 78, 1047–1052. [Google Scholar] [CrossRef]
- Jiang, F.; Herman, G.E. Analysis of Nsdhl-deficient embryos reveals a role for Hedgehog signaling in early placental development. Hum. Mol. Genet. 2006, 15, 3293–3305. [Google Scholar] [CrossRef] [Green Version]
- Katayama, S.; Ashizawa, K.; Gohma, H.; Fukuhara, T.; Narumi, K.; Tsuzuki, Y.; Tatemoto, H.; Nakada, T.; Nagai, K. The expression of Hedgehog genes (Ihh, Dhh) and Hedgehog target genes (Ptc1, Gli1, Coup-TfII) is affected by estrogenic stimuli in the uterus of immature female rats. Toxicol. Appl. Pharmacol. 2006, 217, 375–383. [Google Scholar] [CrossRef]
- Geisert, R.D.; Whyte, J.J.; Meyer, A.E.; Mathew, D.J.; Juarez, M.R.; Lucy, M.C.; Prather, R.S.; Spencer, T.E. Rapid conceptus elongation in the pig: An interleukin 1 beta 2 and estrogen-regulated phenomenon. Mol. Reprod. Dev. 2017, 84, 760–774. [Google Scholar] [CrossRef] [Green Version]
- Knapczyk-Stwora, K.; Durlej, M.; Duda, M.; Czernichowska-Ferreira, K.; Tabecka-Lonczynska, A.; Slomczynska, M. Expression of oestrogen receptor alpha and oestrogen receptor beta in the uterus of the pregnant swine. Reprod. Domest. Anim. 2011, 46, 1–7. [Google Scholar] [CrossRef] [PubMed]
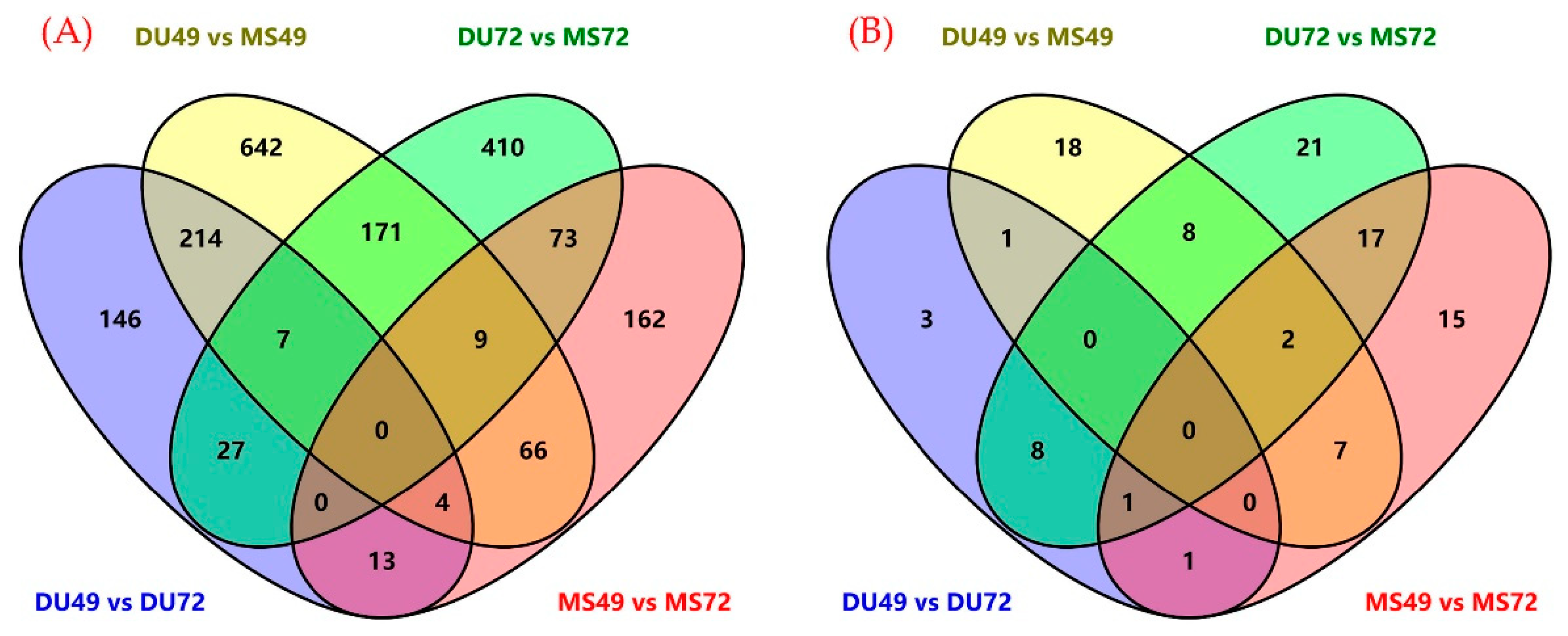
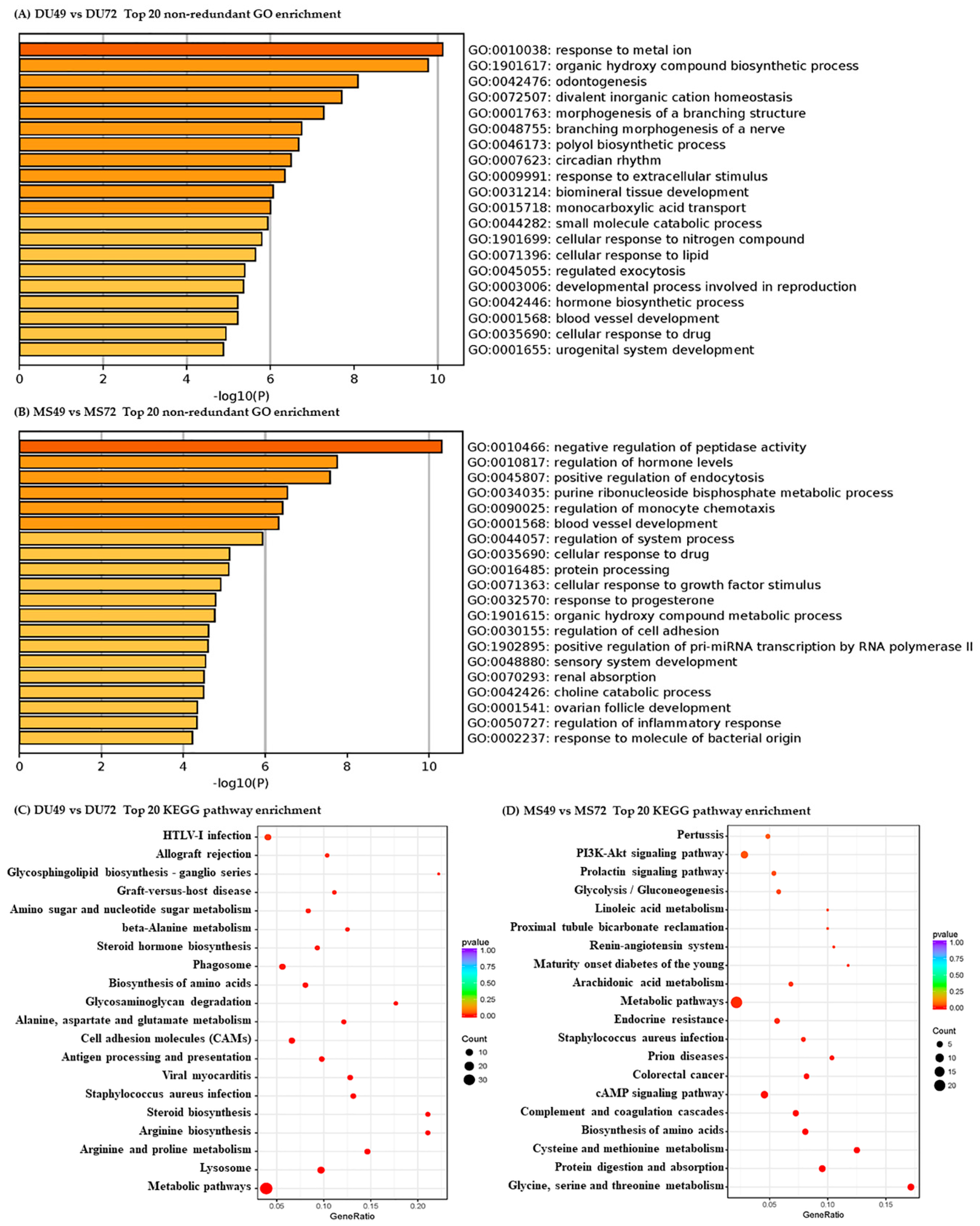
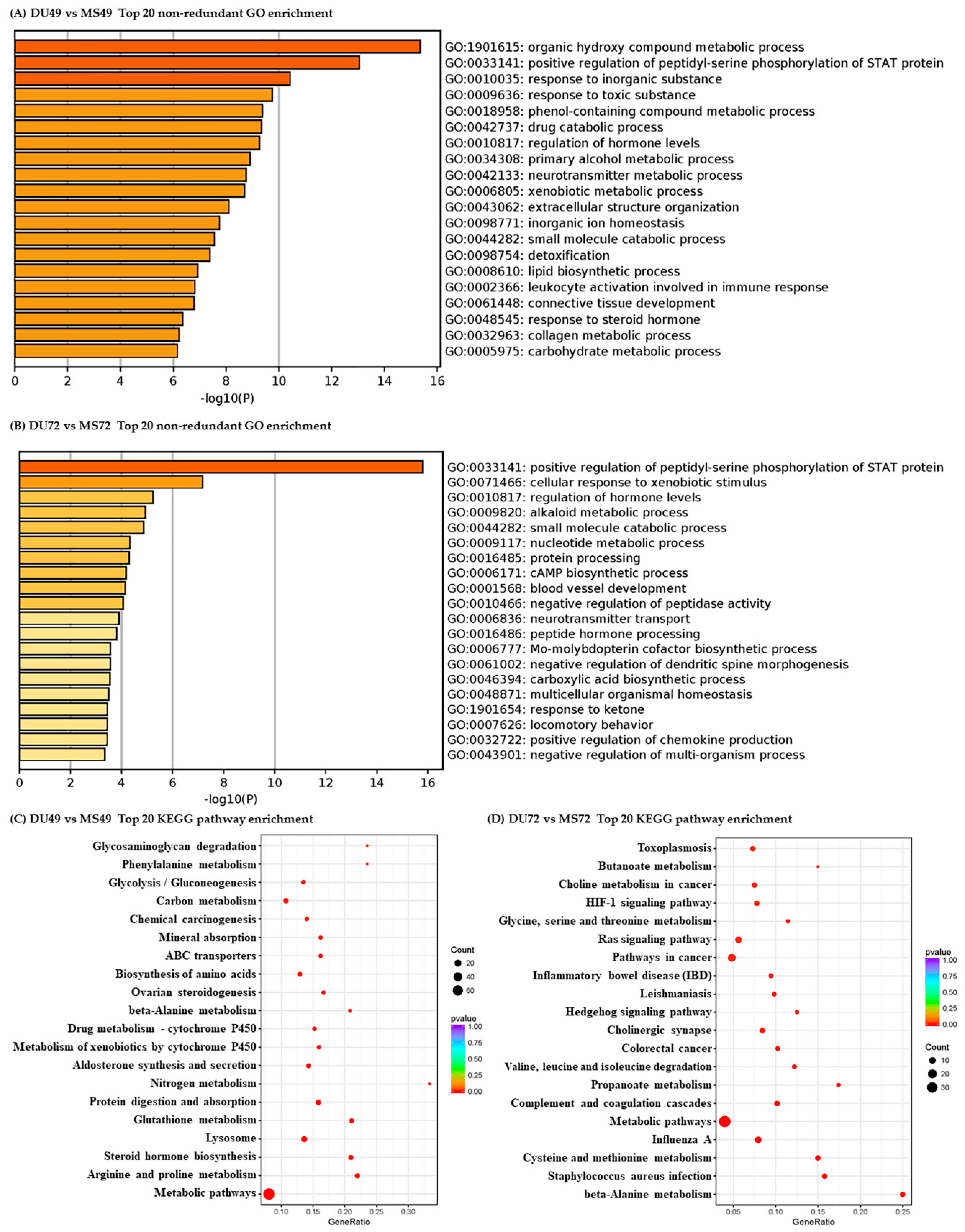
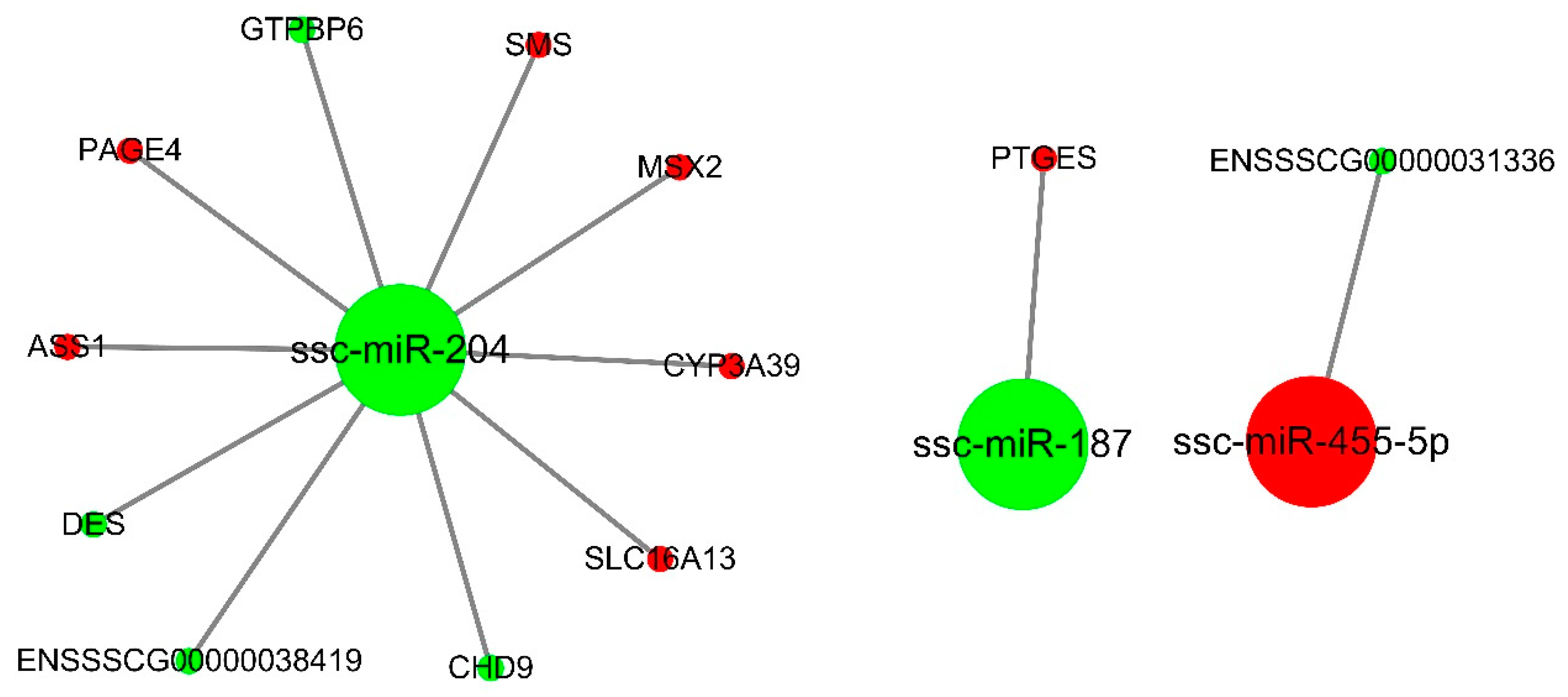

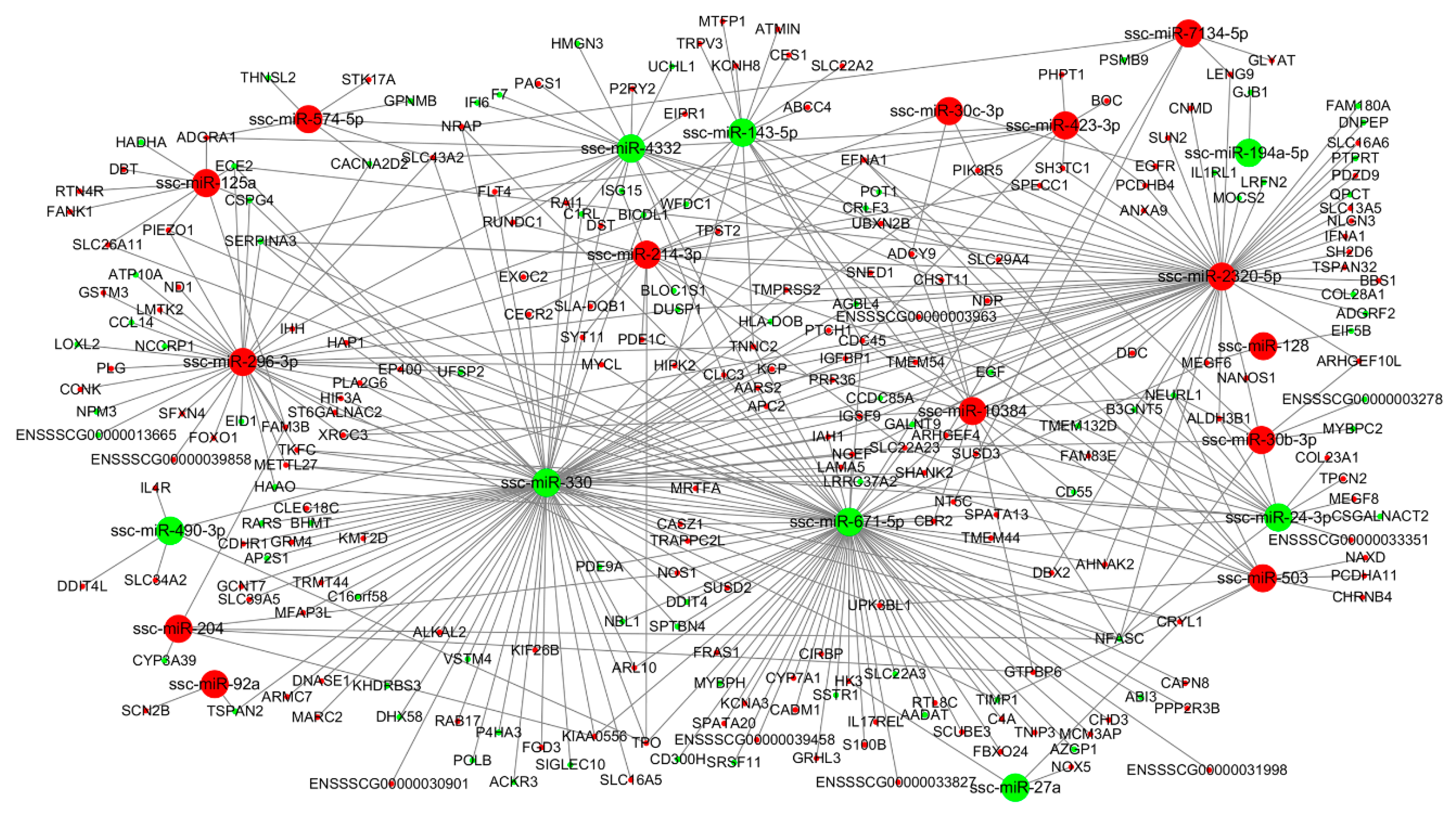

| Comparison | DE mRNAs | DE miRNAs | ||||
|---|---|---|---|---|---|---|
| Total | Up | Down | Total | Up | Down | |
| DU49 vs. DU72 | 411 | 273 | 138 | 14 | 10 | 4 |
| DU49 vs. MS49 | 1113 | 678 | 435 | 36 | 24 | 8 |
| DU72 vs. MS72 | 697 | 397 | 300 | 57 | 26 | 31 |
| MS49 vs. MS72 | 327 | 185 | 142 | 43 | 21 | 22 |
| Comparison Group | Ensembl ID | Gene Name | Log2 Fold Change | Adjusted p-Value |
|---|---|---|---|---|
| DU49 vs. DU72 | ENSSSCG00000030484 | AHR [34] | −0.62 | 0.0247 |
| ENSSSCG00000004283 | DPPA5 [35] | 4.68 | 0.0098 | |
| ENSSSCG00000010698 | FGFR2 [36] | 0.96 | 0.0446 | |
| ENSSSCG00000012229 | GP91-PHOX [37] | −0.74 | 0.0467 | |
| ENSSSCG00000015579 | PTGS2 [38] | 1.85 | 0.0031 | |
| ENSSSCG00000037754 | SLC39A11 [39] | 1.08 | 0.0235 | |
| DU49 vs. MS49 | ENSSSCG00000013303 | ABTB2 [40] | −0.98 | 0.0497 |
| ENSSSCG00000017101 | ADCY2 [41] | −1.35 | 0.0137 | |
| ENSSSCG00000011437 | ALAS1 [42] | 1.33 | 0.0283 | |
| ENSSSCG00000025578 | ALDH1A2 [43] | −1.24 | 0.0422 | |
| ENSSSCG00000004283 | DPPA5 | 3.85 | 3.66 × 10−6 | |
| ENSSSCG00000012229 | GP91-PHOX | −1.05 | 0.0174 | |
| ENSSSCG00000016204 | IHH [44] | −1.50 | 0.0049 | |
| ENSSSCG00000040575 | ISG15 [45] | −1.23 | 0.0171 | |
| ENSSSCG00000015579 | PTGS2 | 1.87 | 0.0419 | |
| ENSSSCG00000007836 | SCNN1G [38] | 1.44 | 0.0412 | |
| DU72 vs. MS72 | ENSSSCG00000009134 | EGF [46] | −1.57 | 0.0012 |
| ENSSSCG00000014336 | EGR1 [47] | −1.44 | 3.75 × 10−6 | |
| ENSSSCG00000002383 | FOS [47] | −0.79 | 0.0040 | |
| ENSSSCG00000010639 | HABP2 [41] | 2.48 | 0.0248 | |
| ENSSSCG00000016204 | IHH | 2.20 | 5.36 × 10−8 | |
| ENSSSCG00000040575 | ISG15 | −1.01 | 0.0059 | |
| ENSSSCG00000000455 | LRIG3 [48] | 2.14 | 1.37 × 10−10 | |
| MS49 vs. MS72 | ENSSSCG00000014336 | EGR1 | −1.59 | 0.0002 |
| ENSSSCG00000025777 | ESR1 [49] | 0.85 | 0.0036 | |
| ENSSSCG00000002383 | FOS | −1.34 | 9.03 × 10−8 | |
| ENSSSCG00000015144 | GRAMD1B [40] | −1.39 | 0.0294 | |
| ENSSSCG00000016204 | IHH | 4.13 | 6.46 × 10−19 |
| Comparison Group | Terms | DEGs No. | p-Value | Genes |
|---|---|---|---|---|
| DU72 vs. MS72 | GO:0032355 ~ Response to estradiol | 7 | 0.00025 | DUSP1, EGFR, F7, IHH, PTCH1, SSTR1, WFDC1 |
| GO:0070482 ~ Response to oxygen levels | 10 | 0.00379 | F7, FOXO1, LOXL2, NOS1, POLB, PSMB9, S100B, HIPK2, DDIT4, HIF3A | |
| GO:0001525 ~ Angiogenesis | 13 | 0.00337 | CSPG4, EFNA1, EGF, FLT4, IHH, LOXL2, GPNMB, CNMD, HIPK2, ACKR3, HIF3A, NOX5, COL23A1 | |
| GO:0001568 ~ Blood vessel development | 15 | 0.00497 | CSPG4, EFNA1, EGF, MEGF8, FOXO1, FLT4, IHH, LOXL2, GPNMB, CNMD, HIPK2, ACKR3, HIF3A, NOX5, COL23A1 | |
| ssc04015 ~ Rap1 signaling pathway | 4 | 3.52 × 10−5 | FLT4, EGF, EGFR, EFNA1 | |
| ssc04014 ~ Ras signaling pathway | 4 | 5.41 × 10−5 | FLT4, EGF, EGFR, EFNA1 | |
| ssc04151 ~ PI3K-Akt signaling pathway | 3 | 0.00019 | FLT4, EGF, EGFR, EFNA1 | |
| ssc04068 ~ FoxO signaling pathway | 4 | 0.00031 | EGF, EGFR, FOXO1 | |
| MS49 vs. MS72 | GO:0032355 ~ Response to estradiol | 5 | 0.00037 | BAD, DUSP1, ESR1, IHH, WNT7A |
| GO:0070482 ~ Response to oxygen levels | 8 | 0.00045 | AQP1, BAD, CLDN3, SMAD3, NOS1, PCK1, ANKRD1, HIPK2 | |
| GO:0001525 ~ Angiogenesis | 8 | 0.00550 | AQP1, CDH13, EFNA1, IHH, NGFR, WNT7A, HIPK2, NOX5 | |
| GO:0001568 ~ Blood vessel development | 9 | 0.00809 | AQP1, CDH13, DHCR7, EFNA1, IHH, NGFR, WNT7A, HIPK2, NOX5 | |
| ssc04015 ~ Rap1 signaling pathway | 2 | 0.00580 | EFNA1, NGFR | |
| ssc04014 ~ Ras signaling pathway | 3 | 0.00026 | BAD, EFNA1, NGFR | |
| ssc04151 ~ PI3K-Akt signaling pathway | 4 | 2.26 × 10−5 | BAD, EFNA1, PCK1, NGFR | |
| ssc04068 ~ FoxO signaling pathway | 2 | 0.00291 | PCK1, SMAD3 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, K.; Wang, J.; Wang, K.; Luo, Y.; Tang, Q.; Liu, X.; Fang, M. Integrated Analysis of miRNA-mRNA Network Reveals Different Regulatory Patterns in the Endometrium of Meishan and Duroc Sows during Mid-Late Gestation. Animals 2020, 10, 420. https://doi.org/10.3390/ani10030420
Yang K, Wang J, Wang K, Luo Y, Tang Q, Liu X, Fang M. Integrated Analysis of miRNA-mRNA Network Reveals Different Regulatory Patterns in the Endometrium of Meishan and Duroc Sows during Mid-Late Gestation. Animals. 2020; 10(3):420. https://doi.org/10.3390/ani10030420
Chicago/Turabian StyleYang, Kaijie, Jue Wang, Kejun Wang, Yabiao Luo, Qiguo Tang, Ximing Liu, and Meiying Fang. 2020. "Integrated Analysis of miRNA-mRNA Network Reveals Different Regulatory Patterns in the Endometrium of Meishan and Duroc Sows during Mid-Late Gestation" Animals 10, no. 3: 420. https://doi.org/10.3390/ani10030420




