Remarkable Cryptic Diversity of Paratylenchus spp. (Nematoda: Tylenchulidae) in Spain †
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Nematode Sampling and Morphological Identification
2.2. Nematode Molecular Characterization
2.3. Phylogenetic Analyses
3. Results
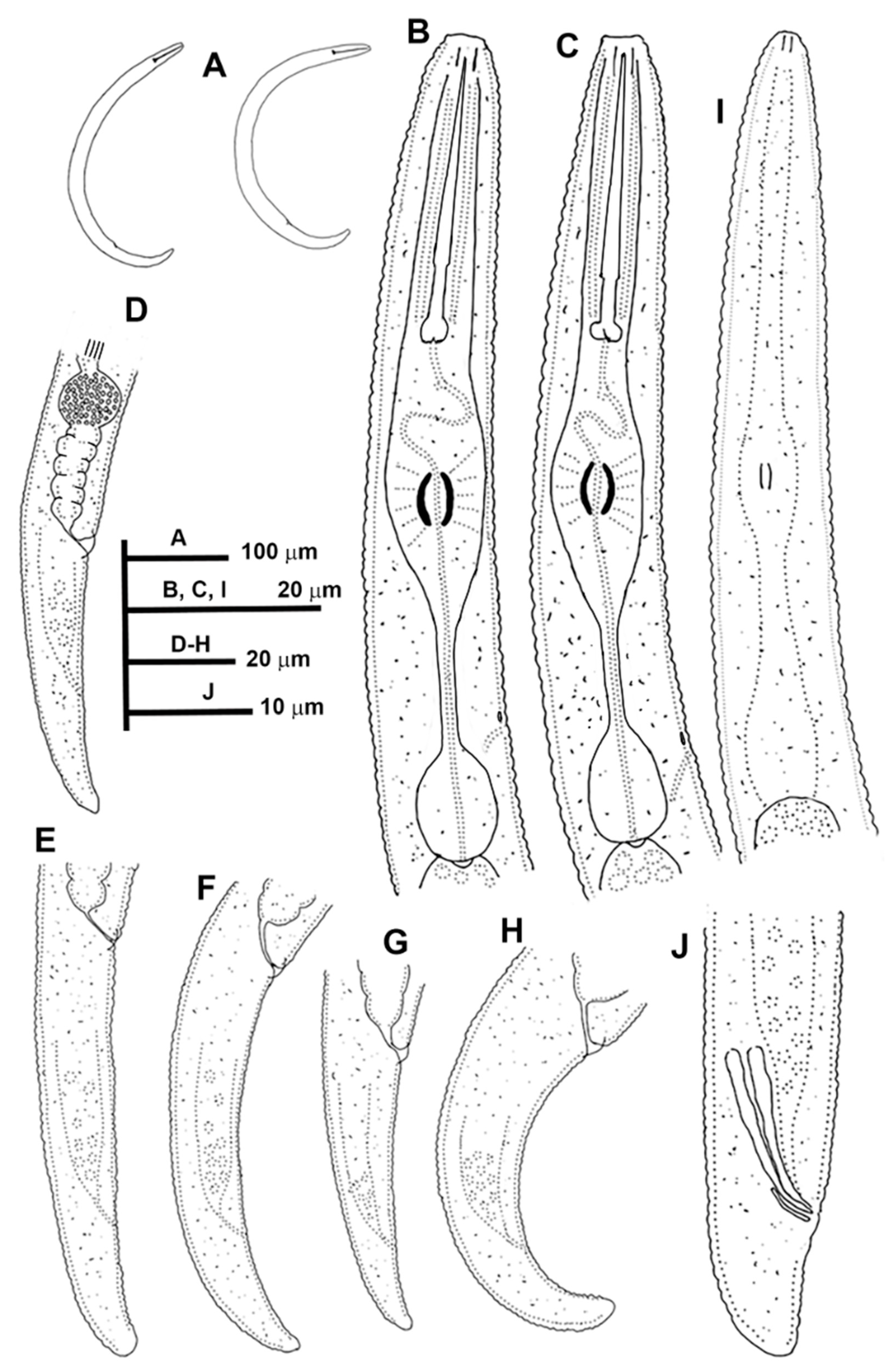
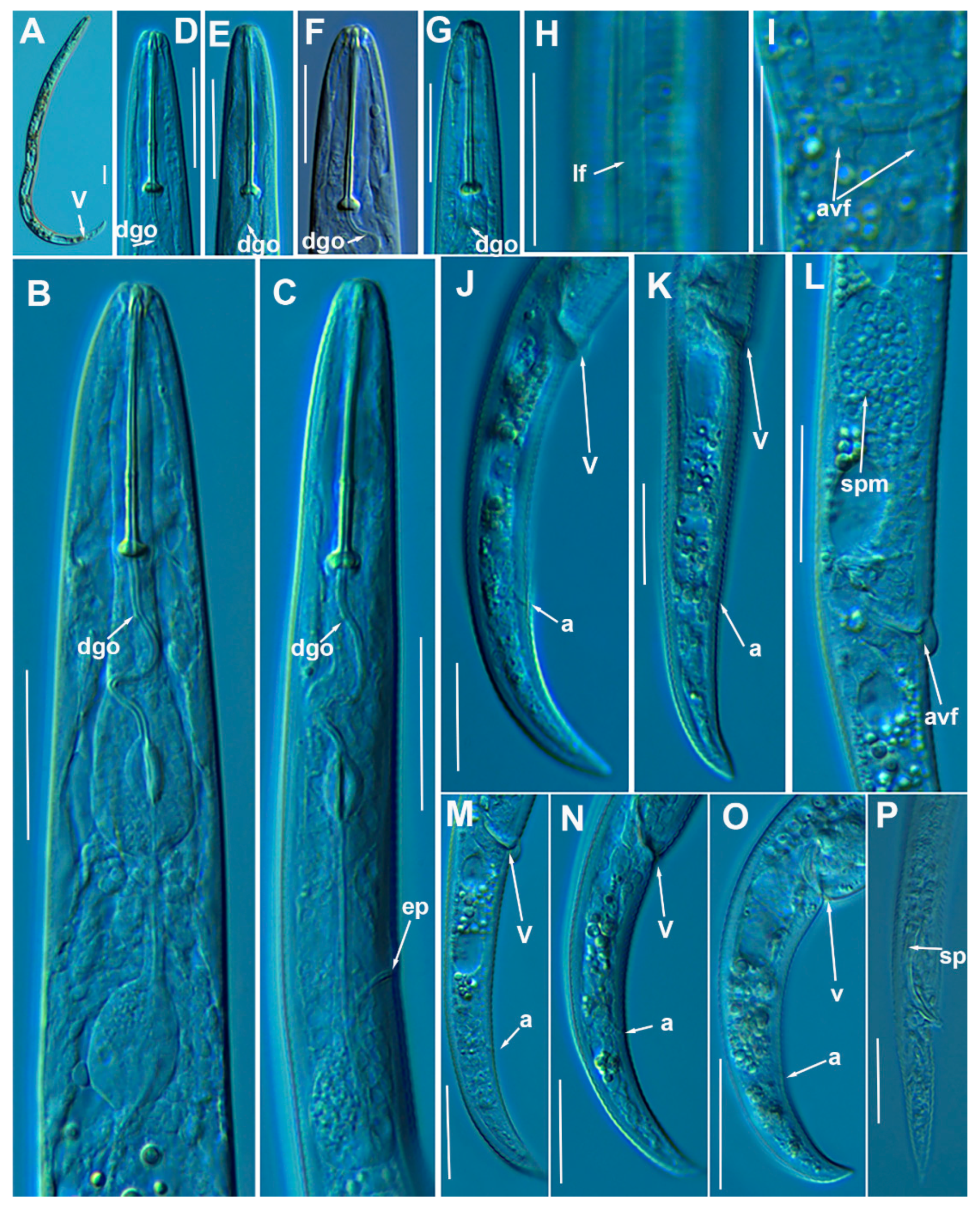
3.1. Systematics
3.1.1. Description of Paratylenchus caravaquenus sp. nov.
Diagnosis and Relationships
Molecular Characterization
Remarks
Type Habitat and Locality
Etymology
Type Material
3.1.2. Description of Paratylenchus indalus sp. nov.
Diagnosis and Relationships
Molecular Characterization
Remarks
Type Habitat and Locality
Etymology
Type Material
3.1.3. Description of Paratylenchus pedrami sp. nov.
Diagnosis and Relationships
Molecular Characterization
Remarks
Etymology
Type Material
3.1.4. Description of Paratylenchus zurgenerus sp. nov.
Diagnosis and Relationships
Molecular Characterization
Remarks
Type Habitat and Locality
Etymology
Type Material
3.1.5. Morphometrics and Remarks of Known Paratylenchus Spanish Populations
3.2. Phylogenetic Analyses
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Micoletzky, H. Die Freilebenden Erd-Nematoden; Archiv für Naturgeschichte: Berlin, Germany, 1922. [Google Scholar]
- Ghaderi, R.; Geraert, E.; Karegar, A. The Tylenchulidae of the World, Identification of the Family Tylenchulidae (Nematoda: Tylenchida); Academia Press: Ghent, Belgium, 2016. [Google Scholar]
- Ghaderi, R. The damage potential of pin nematodes, Paratylenchus Micoletzky, 1922 sensu lato spp. (Nematoda: Tylenchulidae). J. Crop Prot. 2019, 8, 243–257. [Google Scholar]
- Bongers, T.; Ferris, H. Nematode community structure as a bioindicator in environmental monitoring. Trends Ecol. Evol. 1999, 14, 224–228. [Google Scholar] [CrossRef]
- Schmidt, J.H.; Seeger, J.N.; Von Grafenstein, K.; Wintzer, J.; Finckh, M.R.; Hallmann, J. Population dynamics and host range of Paratylenchus bukowinensis. Nematology 2020, 22, 257–267. [Google Scholar] [CrossRef]
- Yeates, G.W.; Lee, W.G. Burning in a New Zealand snow-tussock grassland: Effects on vegetation and soil fauna. N. Z. J. Ecol. 1997, 21, 73–79. [Google Scholar]
- Gaur, H.S. Dissemination and mode of survival of nematodes in dust storms. Indian J. Nematol. 1988, 18, 94–98. [Google Scholar]
- Brzeski, M.W. Paratylenchus Bukowinensis. CIH Descriptions of Plantparasitic Nematodes. Set 6, N 79; Commonwealth Agricultural Bureau: Farnham Royal, UK, 1976. [Google Scholar]
- Brzeski, M.W. Pathogenicity of Paratylenchus bukowinensis Micol. on celery. Rocz. Nauk Rol. Ser. E 1975, 5, 23–29. [Google Scholar]
- Pennacchio, F.; D’Errico, F.P.; Tremblay, E. Spatial distribution pattern and sequential sampling plan for Paratylenchus dianthus. Nematol. Medit. 1985, 13, 137–146. [Google Scholar]
- Allen, M.W.; Jensen, H.J. Cacopaurus epacris, new species (Nematoda: Criconematidae), a nematode parasite of California black walnut roots. Proc. Helm. Soc. Wash. 1950, 17, 10–14. [Google Scholar]
- Thorne, G.; Allen, M.W. Paratylenchus hamatus n.sp, and Xiphinema index n.sp., two nematodes associated with fig roots, with a note on Paratylenchus anceps Cobb. Proc. Helm. Soc. Wash. 1950, 17, 27–35. [Google Scholar]
- French, A.; Lownsbery, B.; Ayoub, S.; Weiner, A.; El-Gholl, N. Pythiaceous fungi and plant-parasitic nematodes in California pear orchards: II. Incidence and distribution of parasitic nematodes in orchard soils. Hilgardia 1964, 35, 603–610. [Google Scholar] [CrossRef] [Green Version]
- Philis, J. Controlling parasitic nematodes in an established vineyard in Cyprus. Nematol. Medit. 2003, 31, 61–63. [Google Scholar]
- Andrássy, I. Paratylenchus microdorus. CIH Descriptions of Plant-Parasitic Nematodes. Set 8, No. 107; Commonwealth Agricultural Bureau: Farnham Royal, UK, 1985. [Google Scholar]
- Odihirin, R.A.; Jenkins, W.R. Host-parasite relationship of impatiens balsamina and certain nematodes. Phytopathology 1965, 55, 736–766. [Google Scholar]
- Braun, A.L.; Lownsbery, B.F. The pin nematode, Paratylenchus neoamblycephalus, on Myrobalan plum and other hosts. J. Nematol. 1975, 7, 336–343. [Google Scholar]
- Webster, G.R.; Orchard, W.R.; Hawn, E.J. Paratylenchus projectus in alfalfa fields of central and northern Alberta. Can. Plant Dis. Surv. 1972, 52, 75–76. [Google Scholar]
- Smolik, J.D. Effects of Paratylenchus projectus on growth of sunflower. Plant Dis. 1987, 71, 975–976. [Google Scholar] [CrossRef]
- Wang, K.; Li, Y.; Xie, H.; Wu, W.-J.; Xu, C.-L. Pin nematode slow decline of Anthurium andraeanum, a new disease caused by the pin nematode Paratylenchus shenzhenensis. Plant Dis. 2016, 100, 940–945. [Google Scholar] [CrossRef] [Green Version]
- Munawar, M.; Yevtushenko, D.P.; Palomares-Rius, J.E.; Castillo, P. Species diversity of pin nematodes (Paratylenchus spp.) from potato growing regions of southern Alberta, Canada. Plants 2021, 10, 188. [Google Scholar] [CrossRef]
- Claerbout, J.; Vandevelde, I.; Venneman, S.; Kigozi, A.; Sutter, N.d.; Neukermans, J.; Bleyaert, P.; Bert, W.; Höfte, M.; Viaene, N. A thorough study of a Paratylenchus sp. in glasshouse-grown lettuce: Characterisation, population dynamics, host plants and damage threshold as keys to its integrated management. Ann. Appl. Biol. 2021, 178, 62–79. [Google Scholar] [CrossRef]
- Van den Berg, E.; Tiedt, L.R.; Subbotin, S.A. Morphological and molecular characterisation of several Paratylenchus Micoletzky, 1922 (Tylenchida: Paratylenchidae) species from South Africa and USA, together with some taxonomic notes. Nematology 2014, 16, 323–358. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.; Karssen, G.; Coureur, M.; Subbotin, S.; Bert, W. Integrative taxonomy and molecular phylogeny of the plant-parasitic nematode genus Paratylenchus (Nematoda: Paratylenchinae): Linking species with molecular barcodes. Plants 2021, 10, 408. [Google Scholar] [CrossRef] [PubMed]
- Fisher, J.M. Studies on Paratylenchus nanus I. effects of variation in environment on several morphometric characters of adults. Nematologica 1965, 11, 269–279. [Google Scholar] [CrossRef]
- Zhuo, K.; Liu, X.; Tao, Y.; Wang, H.; Lin, B.; Liao, J. Morphological and molecular characterisation of three species of Paratylenchus Micoletzky, 1922 (Tylenchida: Paratylenchidae) from China, with a first description of the male P. rostrocaudatus. Nematology 2018, 20, 837–850. [Google Scholar] [CrossRef]
- Mirbabaei, H.; Eskandari, A.; Ghaderi, R.; Karegar, A. On the synonymy of Trophotylenchulus asoensis and T. okamotoi with T. arenarius, and intra-generic structure of Paratylenchus (Nematoda: Tylenchulidae). J. Nematol. 2019, 51, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Munawar, M.; Miao, W.; Castillo, P.; Zheng, J.-W. A new pin nematode, Paratylenchus sinensis n. sp. (Nematoda: Paratylenchinae) in the rhizosphere of white mulberry from Zhejiang Province, China. Eur. J. Plant Pathol. 2020, 156, 1023–1029. [Google Scholar] [CrossRef]
- Subbotin, S.A.; Yan, G.; Kantor, M.; Handoo, Z. On the molecular identity of Paratylenchus nanus Cobb, 1923 (Nematoda: Tylenchida). J. Nematol. 2020, 52, 1–7. [Google Scholar] [CrossRef]
- Sturhan, D.; Geraert, E. Phasmids in Tylenchulidae (Tylenchida: Criconematoidea). Nematology 2005, 7, 249–252. [Google Scholar] [CrossRef]
- Subbotin, S.A.; Vovlas, N.; Crozzoli, R.; Sturhan, D.; Lamberti, F.; Moens, M.; Baldwin, J.G. Phylogeny of Criconematina Siddiqi, 1980 (Nematoda: Tylenchida) based on morphology and D2-D3 expansion segments of the 28S-rRNA gene sequences with application of a secondary structure model. Nematology 2005, 7, 927–944. [Google Scholar] [CrossRef] [Green Version]
- Subbotin, S.A.; Sturhan, D.; Chizhov, V.N.; Vovlas, N.; Baldwin, J.G. Phylogenetic analysis of Tylenchida Thorne, 1949 as inferred from D2 and D3 expansion fragments of the 28S rRNA gene sequences. Nematology 2006, 8, 455–474. [Google Scholar] [CrossRef] [Green Version]
- Brzeski, M.W. Nematodes of Tylenchina in Poland and Temperate Europe; Muzeum i Instytut Zoologii Polska Akademia Nauk: Warsaw, Poland, 1998. [Google Scholar]
- Nguyen, C.N.; Baldwin, J.G.; Choi, Y.E. New records of Paratylenchus Micoletzky, 1922 (Nematoda: Paratylenchinae) from Viet Nam with description of Paratylenchus lapcaiensis sp. n. J. Nematode Morphol. Syst. 2004, 7, 51–75. [Google Scholar]
- Decraemer, W.; Hunt, D.J. Structure and classification. Plant Nematol. 2006, 3–32. [Google Scholar] [CrossRef]
- Ghaderi, R.; Kashi, L.; Karegar, A. Contribution to the study of the genus Paratylenchus Micoletzky, 1922 sensu lato (Nematoda: Tylenchulidae). Zootaxa 2014, 3841, 151–187. [Google Scholar] [CrossRef] [Green Version]
- Hesar, A.M.; Karegar, A.; Ghaderi, R. Phylogenetic relationships of Cacopaurus pestis Thorne, 1943 within representatives of the Tylenchulidae Skarbilovich, 1947 as inferred from ITS and D2-D3 expansion segments of 28S-rRNA sequences. Nematology 2019, 21, 971–994. [Google Scholar] [CrossRef]
- Thorne, G. Cacopaurus pestis, n. g., n. spec. (Nematoda: Criconematinae), a destructive parasite of the walnut, Juglans regia Linn. Proc. Helm. Soc. Wash. 1943, 10, 78–83. [Google Scholar]
- Goodey, T. Soil and Freshwater Nematodes, 2nd ed.; Goodey, J.B., Ed.; Methuen: London, UK, 1963; p. 544. [Google Scholar]
- Siddiqi, M.R. Tylenchida: Parasites of Plants and Insects, 2nd ed.; CABI Publishing: Wallingford, UK, 2000. [Google Scholar]
- Andrássy, I. Free-Living Nematodes of Hungary, II (Nematoda, Errantia); Hungarian Natural History Museum: Budapest, Hungary, 2007. [Google Scholar]
- Brzeski, M.; Hanel, L.; Nico, A.; Castillo, P. Paratylenchinae: Redescription of Paratylenchus arculatus Luc & de Guiran, 1962, a new senior synonym of P. nainianus Edward & Misra, 1963 (Nematoda: Tylenchulidae). Nematology 1999, 1, 375–380. [Google Scholar] [CrossRef]
- Nico, A.I.; Rapoport, H.F.; Jiménez-Díaz, R.M.; Castillo, P. Incidence and population density of plant-parasitic nematodes associated with olive planting stocks at nurseries in southern Spain. Plant Dis. 2002, 86, 1075–1079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peña-Santiago, R. Plant-parasitic nematodes associated with olive (Olea europea L.) in the province of Jaén, Spain. Rev. Nématol. 1990, 13, 113–115. [Google Scholar]
- Gomez-Barcina, A.; Castillo, P.; Pais, M.A.G. Four species of the genus Paratylenchus Micoletzky from Southeasthern Spain. Nematol. Medit. 1990, 18, 169–177. [Google Scholar]
- Talavera, M.; Navas, A. Incidence of plant-parasitic nematodes in natural and semi-natural mountain grassland and the host status of some common grass species. Nematology 2002, 4, 541–552. [Google Scholar] [CrossRef]
- Archidona-Yuste, A.; Wiegand, T.; Castillo, P.; Navas-Cortés, J.A. Dataset on the diversity of plant-parasitic nematodes in cultivated olive trees in southern Spain. Data Brief 2019, 27, 104658. [Google Scholar] [CrossRef]
- Castillo, P.; González-País, M.A.; Gómez-Barcina, A. El género Gracilacus Raski, 1962 en España (Paratylenchinae: Tylenchida). Rev. Ibér. Parasitol. 1989, 49, 321–328. [Google Scholar]
- Talavera, M.; Tobar Jimenez, A. Plant parasitic nematodes from unirrigated fields in Alhama, southeastern Spain. Nematol. Medit. 1997, 25, 73–81. [Google Scholar]
- Castillo, P.; Gómez-Barcina, A. Some species of Tylenchida from natural habitats in southeastern Spain. Nematol. Medit. 1988, 16, 75–86. [Google Scholar]
- Peña Santiago, R.; Geraert, E. New data on Aorolaimus perscitus (Doucet, 1980) and Gracilacus teres Raski, 1976 (Nematoda: Tylenchida) associated with olive (Olea europea L.) in the province of Jaén, Spain. Nematologica 1990, 36, 408–416. [Google Scholar] [CrossRef]
- Hernandez, M.; Mateo, M.D.; Jordana, R. Estudio comparativo entre grupos tróficos de Nematodos del suelo de cinco bosques de Navarra (tres naturales y dos de repoblación). Actas II Congr. Mund. Vasco. Sec. Biol. Ambient. 1988, 2, 323–335. [Google Scholar]
- Coolen, W.A. Methods for extraction of Meloidogyne spp. and other nematodes from roots and soil. In Root-Knot Nematodes (Meloidogyne Species). Systematics, Biology and Control; Lamberti, F., Taylor, C.E., Eds.; Academic Press: New York, NY, USA, 1979; pp. 317–329. [Google Scholar]
- Seinhorst, J.W. Killing nematodes for taxonomic study with hot F.A. 4:1. Nematologica 12, 178. Nematologica 1966, 12, 178. [Google Scholar] [CrossRef]
- De Grisse, A.T. Redescription ou modifications de quelques techniques utilisées dans l’étude de nématodes phytoparasitaires. Meded. Rijksfac. Landbouwwet. Gent 1969, 34, 315–359. [Google Scholar]
- Hunt, D.J.; Palomares-Rius, J.E. General morphology and morphometries of plant-parasitic nematodes. In Practical Plant Nematology; Biblioteca Basica de Agricultura: Montecillo, Mexico, 2012; pp. 25–64. [Google Scholar]
- Palomares-Rius, J.E.; Clavero-Camacho, I.; Archidona-Yuste, A.; Cantalapiedra-Navarrete, C.; León-Ropero, G.; Braun Miyara, S.; Karssen, G.; Castillo, P. Global distribution of the reniform nematode genus Rotylenchulus with the synonymy of Rotylenchulus macrosoma with Rotylenchulus borealis. Plants 2021, 10, 7. [Google Scholar] [CrossRef]
- De Ley, P.; Felix, M.A.; Frisse, L.; Nadler, S.; Sternberg, P.; Thomas, W.K. Molecular and morphological characterisation of two reproductively isolated species with mirror-image anatomy (Nematoda: Cephalobidae). Nematology 1999, 1, 591–612. [Google Scholar] [CrossRef]
- Subbotin, S.A.; Vierstraete, A.; De Ley, P.; Rowe, J.; Waeyenberge, L.; Moens, M.; Vanfleteren, J.R. Phylogenetic relationships within the cyst-forming nematodes (Nematoda, Heteroderidae) based on analysis of sequences from the ITS regions of ribosomal DNA. Mol. Phylogenet. Evol. 2001, 21, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Bowles, J.; Blair, D.; McManus, D.P. Genetic variants within the genus Echinococcus identified by mitochondrial DNA sequencing. Mol. Biochem. Parasitol. 1992, 54, 165–173. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, T.A. BioEdit: A user-friendlybiological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [Green Version]
- Rambaut, A. FigTree v1.4.2, A Graphical Viewer of Phylogenetic Trees. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 27 March 2021).
- Oostenbrink, M. A note on Paratylenchus in the Netherlands with the description of P. goodeyi n. sp. (Nematoda, Criconematidae). Tijdschr. Plantenziekten 1953, 59, 207–216. [Google Scholar] [CrossRef]
- Geraert, E. The Genus Paratylenchus. Nematologica 1965, 11, 301–334. [Google Scholar] [CrossRef]
- Sturhan, D. Plant-parasitic nematodes in Germany—An annotated checklist. Soil Org. 2014, 83, 22. [Google Scholar]
- Izatullaeva, R.I. Nematodes of the Flowering Plants of Kazakhstan; Abstract Kandidat’s Dissertation: Alma-Ata, Kazakhstan, 1967; pp. 1–18. [Google Scholar]
- Lisetskaya, L.F. Nematode fauna of ethereal oil plants of Moldavia. Mater. Nats. Konf. Po Parazitol. Sofia 1968, 13, 47–48. [Google Scholar]
- Solov’eva, G.I. Parasitic Nematodes of Woody and Herbaceous Plants. A Review of the Genus Paratylenchus Micoletzky, 1922 (Nematoda: Criconematidae); Amerind Publishing Co.: New Delhi, India, 1975; p. 134. [Google Scholar]
- Szczygiel, A. Plant parasitic nematodes associated with strawberry plants in Poland. Zesz. Probl. Postepów Nauk Rol. 1974, 154, 1–132. [Google Scholar]
- Brzeski, M.W. Paratylenchinae: Morphology of Some Known Species and Descriptions of Gracilacus Bilineata Sp. N. and G. Vera Sp. N. (Nematoda: Tylenchulidae). Nematologica 1995, 41, 535–565. [Google Scholar] [CrossRef]
- Háněl, L.; Čerevková, A. Diversity of soil nematodes in meadows of the White Carpathians. Helminthologia 2006, 43, 109–116. [Google Scholar] [CrossRef] [Green Version]
- Raski, D.J. Revision of the genus Paratylenchus Micoletzky, 1922 and descriptions of new species. Part II of three parts. J. Nematol. 1975, 7, 274–295. [Google Scholar]
- Powers, T.O.; Harris, T.S.; Higgins, R.S.; Mullin, P.G.; Powers, K.S. Nematode biodiversity assessments need vouchered databases: A BOLD reference library for plant-parasitic nematodes in the superfamily Criconematoidea. Genome 2020, 64, 232–241. [Google Scholar] [CrossRef] [PubMed]
- Raski, D.J. Paratylenchoides gen. n. and two new Species (Nematoda: Paratyleiichidae). Proc. Helm. Soc. Wash. 1973, 40, 230–233. [Google Scholar]
- Myers, R.F. Plant Parasitic Nematodes in New Jersey. J. Nematol. 1986, 18, 272–274. [Google Scholar] [PubMed]
- Esmaeili, M.; Heydari, R.; Castillo, P.; Bidhendi, M.Z.; Palomares-Rius, J.E. Molecular characterisation of two known species of Paratylenchus Micoletzky, 1922 from Iran with notes on the validity of Paratylenchus audriellus Brown, 1959. Nematology 2016, 18, 591–604. [Google Scholar] [CrossRef]
- Wu, L.Y. Paratylenchus veruculatus n. sp. (Criconematidae: Nematoda) from Scotland. Can. J. Zool. 1962, 40, 773–775. [Google Scholar] [CrossRef]
- Raski, D.J. Revision of the genus Paratylenchus Micoletzky, 1922 and descriptions of new species. Part I of three parts. J. Nematol. 1975, 7, 15–34. [Google Scholar]
- Ghaderi, R.; Karegar, A. Some species of Paratylenchus (Nematoda: Tylenchulidae) from Iran. Iran. J. Plant Pathol. 2013, 49, 137–156. [Google Scholar]
- Palomares-Rius, J.E.; Cantalapiedra-Navarrete, C.; Castillo, P. Cryptic species in plant-parasitic nematodes. Nematology 2014, 16, 1105–1118. [Google Scholar] [CrossRef]
- Archidona, Y.; Cai, R.; Cantalapiedra-Navarrete, C.; Carreira, J.A.; Rey, A.; Viñegla, B.; Liébanas, G.; Palomares-Rius, J.E.; Castillo, P. Morphostatic Speciation within the Dagger Nematode Xiphinema hispanum-Complex Species (Nematoda: Longidoridae). Plants 2020, 9, 1649. [Google Scholar] [CrossRef]
- Jörger, K.M.; Norenburg, J.L.; Wilson, N.G.; Schrödl, M. Barcoding against a paradox? Combined molecular species delineations reveal multiple cryptic lineages in elusive meiofaunal sea slugs. BMC Evol. Biol. 2012, 12, 245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palomares-Rius, J.E.; Cantalapiedra-Navarrete, C.; Archidona-Yuste, A.; Subbotin, S.A.; Castillo, P. The utility of mtDNA and rDNA for barcoding and phylogeny of plant-parasitic nematodes from Longidoridae (Nematoda, Enoplea). Sci. Rep. 2017, 7, 10905. [Google Scholar] [CrossRef] [PubMed] [Green Version]




















| Soil Sample Code | Nematode Species | Locality (Province) | Host Plant | D2-D3 | ITS | COI |
|---|---|---|---|---|---|---|
| PI_ARC | Paratylenchus caravaquenus sp. nov. | Caravaca (Murcia) a | pine | MW798270-MW798272 | MW798316-MW798318 | MW797003-MW797004 |
| PR_104 | Paratylenchus indalus sp. nov. | Santa María de Nieva (Almería) a | almond | MW798273-MW798275 | MW798319-MW798322 | MW797005 |
| PR_114 | Paratylenchus indalus sp. nov. | Urracal (Almería) | almond | MW798276-MW798277 | MW798323-MW798324 | MW797006 |
| PR_118 | Paratylenchus indalus sp. nov. | Serón (Almería) | almond | MW798278-MW798280 | MW798325-MW798326 | MW797007 |
| PR_119 | Paratylenchus indalus sp. nov. | El Hijate (Almería) | almond | MW798281-MW798282 | MW798327-MW798328 | MW797008 |
| PR_014 | Paratylenchus pedrami sp. nov. | Córdoba (Córdoba) a | almond | MW798283 | MW798329 | MW797009 |
| PR_017 | Paratylenchus pedrami sp. nov. | Córdoba (Córdoba) | almond | MW798284-MW798285 | MW798330 | - |
| PR_111 | Paratylenchus zurgenerus sp. nov. | Zurgena (Almería) a | almond | MW798286-MW798289 | MW798331-MW798334 | MW797010-MW797011 |
| PR_152 | Paratylenchus baldaccii Raski, 1975 | Cantillana (Sevilla) | peach | MW798290-MW798291 | MW798335-MW798336 | MW797012 |
| PR_193 | Paratylenchus enigmaticus Munawar, 2021 | La Almunia (Zaragoza) | cherry | MW798292 | MW798337 | MW797013 |
| PR_014 | Paratylenchus goodeyi (Oostenbrink, 1953) Raski, 1962 | Córdoba (Córdoba) | almond | MW798293-MW798294 | MW798338-MW798339 | MW797014-MW797015 |
| PR-044 | Paratylenchus hamatus Thorne & Allen, 1950 | Gibraleón (Huelva) | peach | MW798295 | MW798340 | MW797016 |
| PR_115 | Paratylenchus hamatus Thorne & Allen, 1950 | Lúcar (Almería) | almond | MW798296 | MW798341 | - |
| PR_207 | Paratylenchus hamatus Thorne & Allen, 1950 | Sástago (Zaragoza) | peach | MW798297 | - | MW797017 |
| PR_187 | Paratylenchus hamatus Thorne & Allen, 1950 | Ariza (Zaragoza) | almond | MW798298-MW798299 | - | - |
| PR_082 | Paratylenchus holdemani Raski, 1975 | Martos (Jaén) | almond | MW798300 | MW798342 | MW797018 |
| PR_079 | Paratylenchus israelensis (Raski, 1973) Siddiqi, 1986 | Valenzuela (Córdoba) | almond | MW798301-MW798302 | MW798343–MW798346 | - |
| PR_011 | Paratylenchus israelensis (Raski, 1973) Siddiqi, 1986 | Córdoba (Córdoba) | almond | MW798303-MW798305 | - | MW797019-MW797020 |
| PR_124 | Paratylenchus tenuicaudatus Wu, 1961 | Caravaca (Murcia) | almond | MW798306 | MW798347 | MW797021 |
| PR_129 | Paratylenchus tenuicaudatus Wu, 1961 | Calasparra (Murcia) | nectarine | MW798307 | MW798348 | MW797022 |
| PR_168 | Paratylenchus tenuicaudatus Wu, 1961 | Sollana (Valencia) | nectarine | MW798308 | MW798349 | MW797023 |
| PR_208 | Paratylenchus tenuicaudatus Wu, 1961 | Sástago (Zaragoza) | apricot | MW798309 | - | - |
| PR_122 | Paratylenchus veruculatus Wu, 1962 | El Moral (Murcia) | almond | MW798310 | - | MW797026 |
| PR_106 | Paratylenchus veruculatus Wu, 1962 | Santa María de Nieva (Almería) | almond | MW798311-MW798312 | MW798350-MW798351 | MW797027-MW797029 |
| PR_115 | Paratylenchus veruculatus Wu, 1962 | Lúcar (Almería) | almond | MW798313 | MW798352 | MW797024 |
| PR_118 | Paratylenchus veruculatus Wu, 1962 | Serón (Almería) | almond | MW798314 | MW798353 | MW797025 |
| PR_193 | Paratylenchus veruculatus Wu, 1962 | La Almunia (Zaragoza) | cherry | MW798315 | MW798354 | - |
| Measurements and Ratios | Holotype Female | Paratype Females | Paratype Males |
|---|---|---|---|
| Sample code | PI_ARC | PI_ARC | PI_ARC |
| Locality | Caravaca, Murcia | Caravaca, Murcia | Caravaca, Murcia |
| n | 1 | 19 | 6 |
| L | 384 | 384.8 ± 24.4 (344–443) | 419 ± 26.0 (372.5–451.5) |
| a * | 24.8 | 22.6 ± 2.3 (18.4–27.0) | 29.2 ± 2.7 (26.6–33.4) |
| b | 3.5 | 3.7 ± 0.4 (3.2–4.9) | 4.4 ± 0.7 (3.7–5.1) |
| c | 15.7 | 13.9 ± 1.0 (11.2–16.2) | 10.96 ± 0.5 (10.5–11.7) |
| c’ | 2.7 | 2.8 ± 0.2 (2.5–3.2) | 3.6 ± 0.2 (3.3–3.9) |
| V or T | 83.6 | 83.4 ± 0.7 (81.8–84.5) | 49.5 |
| G1 | 33.6 | 36.0 ± 4.5 (29.7–49.0) | - |
| Stylet length | 28.0 | 29.8 ± 1.5 (26.5–32) | - |
| Conus length | 18.0 | 19.5 ± 1.5 (16.5–21) | - |
| m | 64.3 | 65.3 ± 2.6 (60.0–70.0) | - |
| DGO | 7.5 | 6.8 ± 0.8 (5.5–8) | - |
| O | 26.8 | 22.9 ± 2.9 (17.2–30.2) | - |
| Lip width | 6.0 | 6.1 ± 0.5 (5.5–7.0) | 4.7 ± 0.3 (4.5–5.0) |
| Median bulb length | 23.0 | 22.1 ± 3.5 (18.0–29.0) | - |
| Median bulb width | 9.0 | 10.1 ± 1.1 (9.0–13.5) | - |
| Anterior end to center median bulb | 64.5 | 60.5 ± 5.5 (47.0–72.0) | 50.5 |
| MB | 58.6 | 57.9 ± 2.9 (52.2–64.4) | - |
| Nerve ring to anterior end | 84.0 | 80.9 ± 7.8 (65.0–96.0) | - |
| Excretory pore to anterior end | 90.0 | 86.9 ± 4.8 (77.0–98.0) | 73.0 |
| Pharynx length | 110.0 | 104.7 ± 8.6 (89.0–124.0) | 94.8 ± 5.8 (88.8–100) |
| Maximum body diam. | 15.5 | 17.2 ± 1.9 (14.5–21.0) | 14.5 ± 1.2 (13.5–16.0) |
| Tail length | 24.5 | 27.9 ± 2.3 (25.0–32.0) | 38.3 ± 1.9 (35.5–40.5) |
| Anal body diam. | 9.0 | 10.1 ± 0.6 (9.0–11.0) | 10.7 ± 0.8 (9.5–11.5) |
| Spicules | - | - | 23.3 ± 0.5 (22.5–24.0) |
| Gubernaculum | - | - | 5.3 ± 0.4 (5.0–6.0) |
| Measurements and Ratios | Holotype Female | Paratype Females | Females | Females | Females |
|---|---|---|---|---|---|
| Sample code | PR_104 | PR_104 | PR_114 | PR_118 | PR_119 |
| Locality | Sta. Mª Nieva, Almería | Sta. Mª Nieva, Almería | Urracal, Almería | Serón, Almería | El Hijate, Almería |
| n | 1 | 13 | 4 | 2 | 3 |
| L | 377 | 377.8 ± 36.3 (317–434) | 450.5 ± 47.9 (382–484) | 370, 399 | 385 ± 26.2 (357–409) |
| a * | 22.2 | 19.4 ± 2.3 (16.0–23.7) | 24.0 ± 2.3 (21.2–26.8) | 19.5, 20.6 | 20.7 ± 1.9 (19.5–22.9) |
| b | 3.9 | 3.7 ± 0.3 (3.3–4.2) | 4.5 ± 0.3 (4.1–4.7) | 3.6, 3.8 | 3.7 ± 0.2 (3.4–3.9) |
| c | 12.6 | 12.0 ± 1.6 (9.0–14.8) | 16.0 ± 1.9 (13.2–17.3) | 13.0, 13.8 | 12.5 ± 1.2 (11.2–13.4) |
| c’ | 3.0 | 3.0 ± 0.1 (2.7–3.1) | 3.0 ± 0.2 (2.8–3.2) | 2.8, 2.9 | 3.0 ± 0.2 (2.9–3.2) |
| V | 83.0 | 82.9 ± 1.4 (80.4–84.5) | 81.6 ± 1.1 (80.6–83.0) | 82.4, 82.7 | 83.4 ± 0.6 (83.0–84.1) |
| G1 | 20.4 | 25.0 ± 4.9 (20.2–38.8) | 33.5 ± 7.6 (28.3–44.5) | 37.0, 39.8 | 29.2 ± 7.7 (24.6–38.0) |
| Stylet length | 30.0 | 28.3 ± 0.9 (26.0–29.5) | 31.0 ± 1.4 (29.0–32.0) | 28.5, 29.0 | 28.8 ± 1.0 (28.0–30.0) |
| Conus length | 19.0 | 18.3 ± 0.6 (17.0–19.0) | 18.1 ± 0.6 (17.5–19.0) | 17.5, 19.0 | 18.7 ± 1.2 (18.0–20.0) |
| m | 63.3 | 64.5 ± 1.6 (62.1–66.7) | 58.6 ± 4.8 (54.7–65.5) | 61.4, 65.5 | 64.7 ± 1.8 (63.2–66.7) |
| DGO | 6.5 | 6.0 ± 0.4 (5.5–6.5) | 6.3 ± 0.3 (6.0–6.5) | 6.5, 6.5 | 5.5 ± 0.0 (5.5–5.5) |
| O | 21.7 | 21.1 ± 1.7 (18.6–23.2) | 20.2 ± 1.6 (18.8–22.4) | 22.4, 22.8 | 19.5 ± 0.2 (19.3–19.6) |
| Lip width | 6.0 | 6.2 ± 0.3 (5.5–6.5) | 6.5 ± 0.4 (6.0–7.0) | 6.5, 7.0 | 6.5 ± 0.5 (6.0–7.0) |
| Median bulb length | 21.0 | 20.7 ± 2.0 (18.0–24.0) | - | 28.0 | - |
| Median bulb width | 11.0 | 10.0 ± 1.0 (9.0–12.0) | - | 12.0 | - |
| Anterior end to center median bulb | 60.0 | 56.4 ± 3.5 (51–65) | 55.0 ± 0.8 (54.0–56.0) | 55.0, 56.0 | 58.3 ± 1.5 (57.0–60.0) |
| MB | 62.5 | 55.2 ± 2.3 (51.2–59.0) | 54.7 ± 2.6 (53.3–58.5) | 53.3, 53.4 | 55.4 ± 1.7 (54.3–57.4) |
| Nerve ring to anterior end | 72.0 | 76.7 ± 6.4 (68.0–92.0) | 70.5 ± 1.3 (69.0–72.0) | 71.0, 72.0 | 76.0 ± 7.0 (71.0–84.0) |
| Excretory pore to anterior end | 86.0 | 91.1 ± 11.0 (79.0–118.0) | 87.0 ± 2.6 (84.0–90.0) | 86.0, 90.0 | 91.3 ± 8.4 (86.0–101.0) |
| Pharynx length | 96.0 | 101.5 ± 9.8 (89.0–127.0) | 100.8 ± 4.8 (94.0–105.0) | 103.0, 105.0 | 105.3 ± 4.5 (101.1–110.0) |
| Maximum body diam. | 17.0 | 19.7 ± 3.0 (14.0–24.0) | 18.8 ± 1.0 (18.0–20.0) | 18.0, 20.5 | 18.7 ± 2.1 (17.0–21.0) |
| Tail length | 30.0 | 32.0 ± 5.3 (27.0–47.0) | 28.3 ± 1.0 (27.0–29.0) | 28.5, 29.0 | 31.0 ± 1.7 (29.0–32.0) |
| Anal body diam. | 10.0 | 10.8 ± 1.5 (9.5–15.0) | 9.5 ± 0.7 (9.0–10.5) | 10.0, 10.5 | 10.3 ± 0.6 (10.0–11.0) |
| Measurements and Ratios | Holotype Female | Paratype Females | Paratype Males | Females |
|---|---|---|---|---|
| Sample code | PR_14 | PR_14 | PR_14 | PR_17 |
| Locality | Córdoba, Córdoba | Córdoba, Córdoba | Córdoba, Córdoba | Córdoba, Córdoba |
| n | 1 | 20 | 3 | 4 |
| L | 295 | 297 ± 30.0 (231–374) | 269.7 ± 21.6 (245–285) | 305 ± 7.7 (294–312) |
| a * | 21.1 | 18.6 ± 3.1 (13.7–23.0) | 22.6 ± 1.7 (21.1–24.5) | 20.1 ± 1.3 (18.9–22.0) |
| b | 3.7 | 3.7 ± 0.3 (3.2–4.5) | 4.5 ± 0.4 (4.0–4.8) | 3.6 ± 0.1 (3.5–3.7) |
| c | 13.1 | 13.0 ± 1.9 (10.0–16.5) | 24.2 ± 1.8 (22.3–25.9) | 8.6 ± 0.6 (8.2–9.6) |
| c’ | 2.6 | 2.8 ± 0.1 (2.6–3.1) | 1.4 ± 0.1 (1.3–1.6) | 3.5 ± 0.1 (3.4–3.7) |
| V or T | 81.4 | 79.9 ± 1.6 (76.0–82.0) | 34.7 ± 4.6 (30.1–39.3) | 81.2 ± 1.0 (80.1–82.3) |
| G1 | 34.9 | 35.8 ± 6.1 (26.2–50.0) | - | 34.4 ± 7.8 (28.8–39.9) |
| Stylet length | 28.0 | 27.8 ± 1.1 (26.0–30.0) | - | 30.9 ± 1.0 (29.5–32.0) |
| Conus length | 22.0 | 18.5 ± 1.2 (16.0–20.5) | - | 22.0 ± 1.6 (20.0–24.0) |
| m | 78.6 | 66.4 ± 2.8 (61.5–70.4) | - | 71.2 ± 3.0 (67.8–75.0) |
| DGO | 4.0 | 3.9 ± 0.3 (3.5–5.0) | - | 4.6 ± 0.5 (4.0–5.0) |
| O | 14.3 | 14.2 ± 1.4 (12.3–19.2) | - | 15.0 ± 1.7 (12.5–16.1) |
| Lip width | 4.5 | 3.8 ± 0.5 (3.0–5.0) | 3.0 ± 0.0 | 5.5 ± 0.4 (5.0–6.0) |
| Median bulb length | 16.5 | 19.2 ± 1.4 (17.0–22.0) | - | 19.4 ± 1.3 (18.0–21.0) |
| Median bulb width | 7.0 | 8.5 ± 0.4 (8.0–9.0) | - | 8.1 ± 0.9 (7.5–9.5) |
| Anterior end to center median bulb | 45.0 | 46.0 ± 2.8 (39.0–50.0) | - | 49.0 ± 0.0 |
| MB | 57.0 | 57.0 ± 2.9 (53.2–65.3) | - | 57.8 ± 0.3 (57.6–58.3) |
| Nerve ring to anterior end | 60.0 | 59.6 ± 3.5 (53.0–65.0) | - | 66.3 ± 1.5 (65.0–68.0) |
| Excretory pore to anterior end | 69.0 | 71.6 ± 5.3 (62.0–80.0) | 65.5 | 77.8 ± 3.5 (74.0–82.0) |
| Pharynx length | 79.0 | 80.1 ± 4.6 (70.0–86.0) | 60.3 ± 9.7 (52.0–71.0) | 84.8 ± 0.5 (84.0–85.0) |
| Maximum body diam. | 14.0 | 16.5 ± 3.3 (12.0–23.0) | 12.0 ± 1.8 (10.0–13.5) | 15.3 ± 1.0 (14.0–16.5) |
| Tail length | 22.5 | 23.2 ± 3.1 (18.0–28.0) | 11.2 ± 1.3 (10.0–12.5) | 35.5 ± 2.6 (32.0–38.0) |
| Anal body diam. | 8.5 | 8.3 ± 0.9 (7.0–9.5) | 7.8 ± 1.0 (7.0–9.0) | 10.1 ± 0.6 (9.5–11.0) |
| Spicules | - | - | 16.2 ± 2.3 (14.0–18.5) | - |
| Gubernaculum | - | - | 3.5 ± 0.5 (3.0–4.0) | - |
| Measurements and Ratios | Holotype Female | Paratype Females |
|---|---|---|
| Sample code | PR_111 | PR_111 |
| n | 1 | 19 |
| L | 370 | 356.9 ± 32.1 (316–418) |
| a * | 20.0 | 20.1 ± 1.7 (17.5–23.7) |
| b | 3.7 | 3.9 ± 0.4 (3.4–4.9) |
| c | 11.9 | 13.8 ± 1.7 (11.0–17.0) |
| c’ | 2.8 | 2.7 ± 0.3 (2.0–3.2) |
| V | 83.2 | 84.8 ± 1.0 (83.3–86.6) |
| G1 | 40.0 | 28.4 ± 6.9 (18.2–43.5) |
| Stylet length | 15.0 | 15.4 ± 0.6 (14.0–16.0) |
| Conus length | 9.0 | 9.0 ± 0.6 (8.0–10.0) |
| m | 60.0 | 58.6 ± 3.4 (53.1–66.7) |
| DGO | 5.5 | 5.0 ± 0.8 (3.5–6.0) |
| O | 36.7 | 32.4 ± 4.4 (22.6–37.5) |
| Lip width | 6.0 | 6.7 ± 0.6 (6.0–8.0) |
| Median bulb length | - | 22.1 ± 1.96 (19.0–26.0) |
| Median bulb width | - | 10.0 ± 0.9 (9.0–12.0) |
| Anterior end to center median bulb | 55.0 | 47.7 ± 3.0 (41.0–52.0) |
| MB | 55.3 | 51.6 ± 2.1 (47.2–55.1) |
| Nerve ring to anterior end | 70.0 | 64.9 ± 4.2 (59.0–73.0) |
| Excretory pore to anterior end | 83.0 | 81.3 ± 7.4 (67.0–94.0) |
| Pharynx length | 99.5 | 92.5 ± 4.5 (85.0–101.0) |
| Maximum body diam. | 18.5 | 17.8 ± 2.0 (15.5–22.0) |
| Tail length | 31.0 | 26.2 ± 3.5 (22.0–34.5) |
| Anal body diam. | 11.0 | 9.7 ± 1.1 (8.0–11.5) |
| Measurements and Ratios | P. baldaccii | P. enigmaticus | P. goodeyi | P. israelensis | |||
|---|---|---|---|---|---|---|---|
| Females | Females | Females | Females | Males | Females | Females | |
| Sample code | PR_152 | PR_193 | PR_14 | PR_82 | PR_82 | PR_79 | PR_11 |
| Locality | Cantillana, Sevilla | La Almunia, Zaragoza | Córdoba, Córdoba | Martos, Jaén | Martos, Jaén | Valenzuela, Córdoba | Córdoba, Córdoba |
| n | 11 | 5 | 8 | 7 | 4 | 7 | 3 |
| L | 309.2 ± 25.0 (272–354) | 364.6 ± 23.5 (324–383) | 411.1 ± 10.5 (396–427) | 396.4 ± 36.5 (345–441) | 420.3 ± 25 (388–441) | 431.6 ± 26.9 (400–464) | 472.7 ± 27.0 (446–500) |
| a * | 20.5 ± 2.7 (14.7–25.3) | 19.9 ± 1.5 (17.6–21.6) | 22.8 ± 2.6 (19.9–26.8) | 25.1 ± 1.4 (23.5–27.2) | 30.5 ± 3.6 (27.5–35.1) | 21.0 ± 2.4 (18.0–23.6) | 25.2 ± 1.4 (23.6–26.2) |
| b | 3.8 ± 0.4 (3.4–4.4) | 3.8 ± 0.2 (3.6–4.2) | 3.9 ± 0.2 (3.5–4.2) | 3.9 ± 0.3 (3.5–4.3) | 4.9 ± 0.24 (4.5–5.1) | 4.1 ± 0.4 (3.8–4.8) | 4.2 ± 0.3 (3.9–4.5) |
| c | 10.0 ± 1.9 (8.0–14.8) | 14.6 ± 1.6 (12.0–16.0) | 11.7 ± 0.6 (10.7–12.2) | 13.8 ± 1.1 (12.2–15.0) | 11.4 ± 1.1 (10.5–13.0) | 11.8 ± 1.6 (9.2–14.4) | 10.5 ± 1.1 (9.4–11.6) |
| c’ | 3.5 ± 0.1 (3.4–3.7) | 2.7 ± 0.1 (2.5–2.8) | 3.6 ± 0.2 (3.2–3.8) | 3.0 ± 0.4 (2.6–3.5) | 3.5 ± 0.22 (3.4–3.9) | 3.4 ± 0.4 (2.9–4.0) | 4.0 ± 0.6 (3.3–4.5) |
| V or T | 79.8 ± 1.0 (77.5–81.2) | 83.4 ± 1.3 (81.2–84.4) | 81.7 ± 0.5 (81.0–82.4) | 81.5 ± 1.1 (79.9–83.4) | 47.5 ± 4.6 (44.8–52.8) | 80.3 ± 1.0 (78.7–81.8) | 78.0 ± 2.0 (76.1–80.0) |
| G1 | 32.7 ± 4.9 (27.0–38.9) | 33.7 ± 7.8 (23.1–42.9) | 30.8 ± 1.9 (29.2–33.9) | 35.9 ± 10.2 (28.7–43.1) | - | 33.0 ± 2.7 (30.0–37.1) | 33.8 ± 3.9 (29.4–36.4) |
| Stylet length | 30.5 ± 1.1 (29.0–33.0) | 27.6 ± 0.9 (27.0–29.0) | 49.6 ± 2.6 (46.0–53.0) | 26.7 ± 1.5 (24.0–29.0) | 14.5 ± 0.6 (14.0–15.0) | 26.0 ± 1.2 (25.0–28.0) | 26.5 ± 0.5 (26.0–27.0) |
| Conus length | 21.2 ± 1.5 (19.0–24.0) | 17.5 ± 0.9 (17.0–19.0) | 40.2 ± 2.6 (37.0–44.0) | 16.5 ± 1.6 (14.5–19.0) | 8.3 ± 0.4 (8.0–8.8) | 16.6 ± 0.7 (15.5–17.5) | 16.5 ± 0.5 (16.0–17.0) |
| m | 69.3 ± 2.9 (63.3–72.7) | 63.4 ± 1.2 (62.5–65.5) | 80.9 ± 1.4 (79.2–83.0) | 61.7 ± 3.2 (57.7–66.7) | 57.5 ± 3.1 (53.3–60.7) | 64.1 ± 2.2 (62.0–68.0) | 62.3 ± 0.7 (61.5–63.0) |
| DGO | 5.5 ± 0.7 (4.5–6.5) | 5.9 ± 1.6 (4.0–7.5) | 5.3 ± 0.4 (4.5–5.5) | 6.4 ± 0.5 (5.5–7.0) | 5.3 ± 0.6 (5.0–6.0) | 6.6 ± 0.7 (6.0–7.5) | 5.8 ± 0.3 (5.5–6.0) |
| O | 17.9 ± 2.5 (15.2–22.4) | 21.5 ± 6.0 (14.8–27.8) | 10.6 ± 1.1 (8.8–12.0) | 24.1 ± 2.0 (21.2–27.1) | 36.3 ± 3.4 (33.3–40.0) | 25.3 ± 2.4 (22.2–28.3) | 22.0 ± 1.2 (20.8–23.1) |
| Lip width | 5.5 ± 0.5 (4.5–6.0) | 7.6 ± 0.4 (7.0–8.0) | 5.1 ± 0.2 (5.0–5.5) | 6.7 ± 0.6 (6.0–7.5) | 3.4 ± 0.5 (3.0–4.0) | 8.8 ± 0.3 (8.5–9.0) | 8.3 ± 0.8 (7.5–9.0) |
| Median bulb length | 19.7 ± 2.5 (17.0–24.0) | 25.0 ± 1.0 (24.0–26.0) | 20.6 ± 2.1 (17.0–23.0) | 20.4 ± 2.8 (16.5–24.0) | 16.7 ± 0.6 (16.0–17.0) | 25.8 ± 3.1 (23.0–31.0) | 24.3 ± 2.1 (22.0–26.0) |
| Median bulb width | 8.6 ± 0.7 (7.5–9.5) | 11.2 ± 0.8 (10.5–12.0) | 10.9 ± 0.3 (10.5–11.5) | 9.8 ± 0.6 (9.0–10.5) | 7.8 ± 0.3 (7.5–8.0) | 11.9 ± 1.2 (10.5–14.0) | 8.5 ± 0.5 (8.0–9.0) |
| Anterior end to center median bulb | 47.6 ± 3.1 (43.0–52.0) | 53.6 ± 3.0 (50.0–57.0) | 73.7 ± 2.8 (70.0–77.0) | 57.1 ± 3.0 (52.5–60.5) | 52.6 ± 5.1 (45.0–56.0) | 60.2 ± 3.8 (55.0–65.0) | 60.7 ± 1.5 (59.0–62.0) |
| MB | 59.0 ± 1.2 (57.6–60.5) | 55.8 ± 1.0 (54.4–56.8) | 68.5 ± 3.4 (62.6–72.8) | 56.4 ± 0.9 (55.2–57.6) | - | 56.8 ± 1.8 (54.7–60.0) | 54.2 ± 0.9 (53.2–54.9) |
| Nerve ring to anterior end | 62.1 ± 4.0 (57.0–68.0) | 70.4 ± 4.0 (66.0–75.0) | 87.5 ± 7.4 (78.0–96.0) | 75.5 ± 5.3 (68.0–85.5) | 62.3 ± 5.3 (58.5–66.0) | 78.4 ± 5.9 (71.0–89.5) | 78.3 ± 3.2 (76.0–82.0) |
| Excretory pore to anterior end | 75.3 ± 6.5 (69.0–90.0) | 87.6 ± 5.7 (81.0–94.0) | 91.6 ± 6.8 (82.0–102.0) | 87.2 ± 6.5 (79.5–96.5) | 79.5 ± 6.1 (72.5–84.0) | 90.2 ± 5.4 (84.0–97.0) | 92.0 ± 1.0 (91.0–93.0) |
| Pharynx length | 82.3 ± 4.5 (75.0–87.0) | 96.2 ± 6.5 (88.0–103.0) | 107 ± 8.3 (97.0–123.0) | 100.7 ± 4.6 (93.0–107.0) | 86.3 ± 5.9 (78.0–91.0) | 106.1 ± 8.3 (95.0–117.0) | 112.0 ± 1.0 (111–113) |
| Maximum body diam. | 15.3 ± 1.6 (12.5–18.5) | 18.4 ± 1.7 (16.5–21.0) | 18.3 ± 2.2 (15.0–20.5) | 15.8 ± 0.8 (14.5–16.5) | 13.9 ± 1.0 (12.5–15.0) | 20.7 ± 2.4 (18.0–23.5) | 18.8 ± 1.6 (17.0–20.0) |
| Tail length | 31.7 ± 5.2 (24.0–40.0) | 25.0 ± 1.4 (23.5–27.0) | 35.3 ± 1.5 (33.5–37.5) | 228.9 ± 3.9 (23.0–33.5) | 37.1 ± 2.7 (34.0–40.5) | 36.9 ± 5.0 (32.0–46.0) | 45.3 ± 4.0 (43.0–50.0) |
| Anal body diam. | 9.0 ± 1.2 (7.0–11.0) | 9.4 ± 0.4 (9.0–10.0) | 9.9 ± 0.6 (9.0–10.5) | 9.8 ± 0.5 (9.0–10.5) | 10.5 ± 0.4 (44.8–52.8) | 11.0 ± 1.1 (10.0–12.5) | 11.5 ± 1.3 (10.5–13.0) |
| Spicules | - | - | - | - | 21.8 ± 0.96 (21.0–23.0) | - | - |
| Gubernaculum | - | - | - | - | 5.1 ± 0.25 (5.0–5.5) | - | - |
| Measurements and Ratios | Females | Females | Females | Females | Males |
|---|---|---|---|---|---|
| Sample code | PR_44 | PR_115 | PR_207 | PR_187 | PR_187 |
| Locality | Gibraleón, Huelva | Lúcar, Almería | Sástago, Zaragoza | Ariza, Zaragoza | Ariza, Zaragoza |
| n | 4 | 4 | 5 | 15 | 2 |
| L | 362 ± 11.0 (347–373) | 386.5 ± 28.8 (355–420) | 421.6 ± 46.9 (377–487) | 373.5 ± 31.4 (327–433.5) | 744, 789 |
| a * | 22.2 ± 1.4 (20.4–23.4) | 20.7 ± 1.8 (19.2–23.3) | 20.5 ± 3.7 (16.5–24.9) | 20.2 ± 3.1 (13.5–25.3) | 40.5, 45.1 |
| b | 4.0 ± 0.2 (3.9–4.2) | 3.8 ± 0.3 (3.4–4.1) | 4.6 ± 0.4 (4.3–5.2) | 3.8 ± 0.2 (3.5–4.4) | 4.7, 6.7 |
| c | 12.9 ± 0.8 (11.7–13.6) | 13.9 ± 1.7 (11.7–15.4) | 13.6 ± 1.8 (11.5–16.2) | 12.6 ± 1.0 (10.4 ± 14.6) | 17.5, 18.6 |
| c’ | 3.3 ± 0.1 (3.2–3.4) | 3.0 ± 0.3 (2.6–3.3) | 3.1 ± 0.3 (2.9–3.6) | 2.9 ± 0.3 (2.3–3.6) | 3.5, 4.3 |
| V or T | 81.7 ± 0.9 (80.4 ± 82.6) | 81.9 ± 0.4 (81.5–82.2) | 81.8 ± 1.3 (80.4–83.6) | 82.0 ± 1.1 (80.1–84.2) | 53.0, 59.1 |
| G1 | 41.7 ± 6.9 (33.3–47.7) | 39.7 ± 4.9 (35.6–45.6) | 55.9 ± 3.2 (52.3–59.9) | 46.5 ± 7.6 (33.0–53.5) | - |
| Stylet length | 31.1 ± 1.9 (29.0–33.0) | 33.4 ± 2.1 (31.5–36.0) | 30.1 ± 0.7 (29.0–31.0) | 30.9 ± 1.1 (28.0–32.5) | 16.5, 19.5 |
| Conus length | 20.8 ± 1.8 (19.0–23.0) | 22.5 ± 1.7 (21.0–25.0) | 20.2 ± 2.2 (18.0–23.0) | 19.8 ± 1.1 (18.0–21.5) | 8.0, 8.0 |
| m | 66.6 ± 2.8 (65.0–70.8) | 67.4 ± 2.6 (64.7–69.8) | 67.0 ± 5.7 (62.1–74.2) | 59.8 ± 2.8 (60.0–69.4) | 41.0, 48.5 |
| DGO | 3.8 ± 0.3 (3.5–4.0) | 7.1 ± 0.9 (6.0–8.0) | 6.5 ± 0.8 (5.5–7.5) | - | 4.5, 5.5 |
| O | 12.0 ± 0.3 (11.7–12.3) | 21.4 ± 3.0 (18.8–25.4) | 21.6 ± 2.4 (18.3–24.2) | - | 27.3, 28.2 |
| Lip width | 4.5 ± 0.4 (4.0–5.0) | 6.3 ± 0.3 (6.0–6.5) | 6.4 ± 0.4 (6.0–7.0) | 6.2 ± 0.4 (5.5–7.0) | 6.5, 7.5 |
| Median bulb length | - | 22.8 ± 2.9 (21.0–27.0) | 26.8 ± 1.0 (26.0–28.0) | 17.9 ± 2.0 (13.5–20.0) | 10.5, 12.5 |
| Median bulb width | - | 9.4 ± 1.9 (7.0–11.5) | 11.0 ± 0.8 (10.0–12.0) | 9.4 ± 1.4 (7.0–12.0) | 8.0, 10.0 |
| Anterior end to center median bulb | 52.5 ± 1.7 (50.0–54.0) | 56.0 ± 1.8 (54.0–58.0) | 55.0 ± 2.0 (53.0–58.0) | 56.1 ± 3.7 (51.0–64.0) | 64.5, 70.0 |
| MB | 58.0 ± 2.2 (55.6–60.2) | 55.4 ± 2.1 (52.9–58.1) | 60.5 ± 3.6 (56.8–65.1) | 57.3 ± 2.6 (53.2–62.2) | 44.6, 55.1 |
| Nerve ring to anterior end | 71.3 ± 5.4 (67.0–79.0) | 74.3 ± 6.9 (67.0–83.0) | 66.0 ± 1.7 (63.0–67.0) | - | 85.5, 87.0 |
| Excretory pore to anterior end | 82.0 ± 5.2 (76.0–88.0) | 85.9 ± 7.1 (78.5–92.0) | 83.8 ± 2.3 (81.0–87.0) | 87.8 ± 6.7 (77.5–100.0) | 104.0, 108.0 |
| Pharynx length | 90.5 ± 3.1 (88.0–95.0) | 101.3 ± 5.6 (93.0–105.0) | 91.0 ± 3.8 (86.0–95.0) | 98.3 ± 6.3 (85.5–109.0) | 117.0, 157.0 |
| Maximum body diam. | 16.4 ± 0.8 (15.5–17.0) | 18.8 ± 2.3 (16.0–21.5) | 21.2 ± 4.9 (18.0–29.0) | 19.0 ± 3.7 (17.0–27.5) | 16.5, 19.5 |
| Tail length | 28.3 ± 2.2 (26.0–31.0) | 28.4 ± 5.5 (23.0–36.0) | 31.2 ± 3.7 (26.0–36.0) | 29.7 ± 2.0 (25.5–33.0) | 40.0, 45.0 |
| Anal body diam. | 8.6 ± 0.5 (8.0–9.0) | 9.5 ± 1.1 (8.5–11.0) | 10.0 ± 0.6 (9.0–10.5) | 10.2 ± 1.2 (8.0–12.5) | 10.5, 11.5 |
| Spicules | - | - | - | - | 22.0, 23.5 |
| Gubernaculum | - | - | - | - | 11.0, 13.5 |
| Measurements and Ratios | Females | Males | Females | Females | Females |
|---|---|---|---|---|---|
| Sample code | PR_124 | PR_124 | PR_129 | PR_168 | PR_208 |
| Locality | Caravaca, Murcia | Caravaca, Murcia | Calasparra, Murcia | Sollana, Valencia | Sástago, Zaragoza |
| n | 3 | 2 | 9 | 4 | 3 |
| L | 368.7 ± 20.4 (354–392) | 356, 364 | 376.6 ± 31.0 (307–407) | 358.5 ± 44.7 (292–389) | 405.3 ± 10.3 (394–414) |
| a * | 23.5 ± 1.3 (22.1–24.5) | 29.1, 29.7 | 22.7 ± 3.7 (16.5–28.4) | 23.6 ± 2.0 (20.9–25.1) | 18.3 ± 0.8 (17.4–18.8) |
| b | 4.5 ± 0.2 (4.2–4.6) | 4.4, 4.7 | 3.8 ± 0.3 (3.4–4.2) | 4.0 ± 0.4 (3.4–4.2) | 4.0 ± 0.0 |
| c | 11.7 ± 1.9 (10.4–13.8) | 11.1, 12.3 | 11.7 ± 1.3 (10.5–14.4) | 9.7 ± 0.9 (8.7–10.5) | 10.3 ± 0.5 (10.0–10.9) |
| c’ | 3.7 ± 0.4 (3.3–4.0) | 3.0, 3.8 | 3.6 ± 0.5 (2.8–4.8) | 3.8 ± 0.5 (3.3–4.2) | 3.4 ± 0.1 (3.3–3.5) |
| V or T | 83.0 ± 1.5 (82.1–84.7) | 39.3, 46.3 | 80.8 ± 1.3 (78.3–82.5) | 80.6 ± 0.8 (79.6–81.5) | 80.2 ± 0.8 (79.2–80.7) |
| G1 | 46.3 ± 10.8 (33.9–52.8) | - | 41.3 ± 7.4 (27.6–50.4) | 34.8 ± 5.5 (29.8–41.2) | 43.7 ± 3.3 (40.1–46.6) |
| Stylet length | 29.3 ± 0.6 (29.0–30.0) | - | 30.7 ± 1.1 (29.0–32.0) | 30.5 ± 1.5 (29.0–32.0) | 32.7 ± 0.8 (32.0–33.5) |
| Conus length | 17.7 ± 0.6 (17.0–18.0) | - | 21.0 ± 1.2 (19.5–23.0) | 20.5 ± 1.9 (18.0–22.0) | 21.7 ± 1.2 (21.0–23.0) |
| m | 60.9 ± 2.0 (58.6–62.1) | - | 68.4 ± 1.7 (66.7–71.9) | 67.1 ± 4.1 (61.0–69.8) | 66.3 ± 2.1 (64.6–68.7) |
| DGO | 5.3 ± 0.6 (5.0–6.0) | - | 5.0 ± 0.9 (4.0–6.5) | 6.7 ± 0.6 (6.0–7.0) | 7.2 ± 0.3 (7.0–7.5) |
| O | 18.4 ± 2.0 (17.2–20.7) | - | 16.3 ± 2.7 (12.7–20.3) | 22.2 ± 1.5 (20.7–23.7) | 21.9 ± 0.4 (21.5–22.4) |
| Lip width | 6.2 ± 0.3 (6.0–6.5) | 4.0 | 4.9 ± 0.8 (4.0–6.5) | 5.3 ± 0.5 (5.0–6.0) | 7.2 ± 0.3 (7.0–7.5) |
| Median bulb length | - | - | 23.4 ± 2.9 (18.0–27.0) | 17.5 ± 2.1 (16.0–19.0) | 28.0 ± 1.0 (27.0–29.0) |
| Median bulb width | - | - | 9.1 ± 1.4 (7.0–11.5) | 8.8 ± 1.3 (7.5–10.0) | 12.8 ± 0.8 (12.0–13.5) |
| Anterior end to center median bulb | 44.7 ± 1.2 (44.0–46.0) | 44.0, 46.0 | 57.9 ± 3.6 (52.0–63.0) | 50.8 ± 3.2 (49.0–54.5) | 61.0 ± 1.0 (60.0–62.0) |
| MB | 54.3 ± 2.4 (51.8–56.4) | 56.1, 58.7 | 57.7 ± 3.4 (54.4–64.2) | 56.2 ± 3.2 (52.7–58.9) | 60.4 ± 0.2 (60.2–60.6) |
| Nerve ring to anterior end | 61.7 ± 4.5 (57.0–66.0) | 54.0, 55.0 | 73.9 ± 5.3 (66.5–81.0) | 64.0 ± 6.2 (57.0–69.0) | 80.3 ± 0.8 (79.2–80.7) |
| Excretory pore to anterior end | 75.3 ± 2.3 (74.0–78.0) | 59.0, 71.0 | 80.4 ± 7.7 (68.0–88.0) | 75.2 ± 2.4 (72.5–77.0) | 93.3 ± 7.2 (85.0–98.0) |
| Pharynx length | 82.3 ± 3.8 (78.0–85.0) | 75.0, 82.0 | 98.1 ± 7.6 (88.0–110.0) | 90.4 ± 3.2 (86.0–93.0) | 101.0 ± 2.0 (99.0–103.0) |
| Maximum body diam. | 15.7 ± 0.6 (15.0–16.0) | 12.0, 12.5 | 17.1 ± 3.8 (12.0–24.0) | 15.1 ± 0.9 (14.0–16.0) | 22.2 ± 1.3 (21.0–23.5) |
| Tail length | 32.0 ± 5.3 (26.0–36.0) | 29.5, 32.0 | 32.6 ± 4.8 (25.0–38.0) | 37.3 ± 6.9 (28.0–43.0) | 39.3 ± 1.5 (38.0–41.0) |
| Anal body diam. | 8.7 ± 0.6 (8.0–9.0) | 8.5, 10.0 | 9.0 ± 1.2 (7.0–11.0) | 9.8 ± 1.7 (8.5–12.0) | 11.7 ± 0.6 (11.0–12.0) |
| Spicules | - | 22.0, 25.0 | - | - | - |
| Gubernaculum | - | 6.0, 7.5 | - | - | - |
| Measurements and Ratios | Females | Females | Females | Females | Females |
|---|---|---|---|---|---|
| Sample code | PR_122 | PR_106 | PR_115 | PR_118 | PR_193 |
| Locality | Puebla de Don Fadrique, Granada | Sta. Mª Nieva, Almería | Lúcar, Almería | Serón, Almería | La Almunia, Zaragoza |
| n | 10 | 4 | 10 | 4 | 10 |
| L | 376.1 ± 30.4 (354–436) | 381.5 ± 25.4 (349–407) | 374.5 ± 37.8 (303–445) | 379.5 ± 19.2 (353–395) | 363.8 ± 45.8 (279–441) |
| a * | 20.4 ± 2.5 (17.0–24.3) | 20.7 ± 0.3 (17.5–24.7) | 20.4 ± 1.9 (16.8–23.9) | 22.2 ± 0.7 (21.4–23.1) | 19.8 ± 1.6 (16.4–21.7) |
| b | 4.0 ± 0.2 (3.7–4.4) | 4.2 ± 0.4 (3.8–4.8) | 4.0 ± 0.4 (3.4–4.7) | 4.1 ± 0.3 (3.8–4.4) | 3.8 ± 0.3 (3.4–4.4) |
| c | 14.8 ± 2.0 (11.8–19.7) | 12.9 ± 0.6 (12.0–13.4) | 15.5 ± 1.7 (13.1–18.4) | 13.7 ± 0.3 (13.5–14.1) | 14.0–1.5 (11.2–15.8) |
| c’ | 2.5 ± 0.2 (2.3–2.8) | 2.7 ± 0.3 (2.3–2.9) | 2.4 ± 0.3 (2.1–2.8) | 2.8 ± 0.04 (2.7–2.8) | 2.8 ± 0.2 (2.5–3.0) |
| V | 84.2 ± 1.0 (82.7–85.5) | 84.0 ± 0.9 (83.1–84.8) | 83.6 ± 0.6 (82.8–84.5) | 83.8 ± 1.1 (82.8–84.9) | 84.3 ± 0.8 (83.1–85.4) |
| G1 | 36.5 ± 7.3 (22.3–47.7) | 34.2 ± 12.5 (26.7–52.9) | 38.5 ± 4.1 (32.0–43.2) | 33.6 ± 0.8 (25.9–43.6) | 35.6 ± 4.7 (29.8–44.4) |
| Stylet length | 15.1 ± 0.6 (14.0–16.0) | 15.9 ± 0.3 (15.5–16.0) | 15.5 ± 0.6 (14.5–16.5) | 15.6 ± 0.25 (15.5–16.0) | 15.7 ± 0.7 (15.0–17.0) |
| Conus length | 9.1 ± 0.7 (7.5–10.0) | 9.9 ± 0.3 (9.5–10.0) | 9.5 ± 0.6 (8.0–10.0) | 9.8 ± 0.3 (9.5–10.0) | 9.6 ± 0.5 (9.0–10.5) |
| m | 60.4 ± 5.5 (50.0–66.7) | 62.2 ± 0.6 (61.3–62.5) | 60.9 ± 2.3 (55.2–63.3) | 61.4 ± 2.5 (59.4–64.5) | 61.3 ± 1.8 (58.1–63.3) |
| DGO | 5.2 ± 1.2 (3.0–7.0) | 4.3 ± 0.3 (4.0–4.5) | 4.5 ± 0.6 (3.5–5.5) | 3.6 ± 0.6 (3.0–4.5) | 4.5 ± 0.4 (4.0–5.0) |
| O | 34.3 ± 7.4 (20.0–46.7) | 26.8 ± 2.1 (25.0–29.0) | 28.7 ± 3.3 (21.9–33.3) | 22.8 ± 3.9 (18.8–28.1) | 28.8 ± 2.7 (25.0–33.3) |
| Lip width | 6.6 ± 0.6 (6.0–7.5) | 6.0 ± 0.4 (5.5–6.5) | 6.3 ± 0.4 (6.0–7.0) | 7.0 ± 0.4 (6.5–7.5) | 6.7 ± 0.4 (6.0–7.5) |
| Median bulb length | 23.8 ± 2.4 (22.0–27.0) | 25.3 ± 1.0 (24.0–26.0) | 24.6 ± 5.0 (19.0–33.0) | 26.5 ± 0.7 (26.0–27.0) | 20.8 ± 1.5 (19.0–22.0) |
| Median bulb width | 8.1 ± 0.6 (7.5–9.0) | 8.4 ± 0.5 (8.0–9.0) | 9.3 ± 1.3 (8.0–12.0) | 9.5 ± 0.7 (9.0–10.0) | 9.3 ± 1.2 (8.0–11.0) |
| Anterior end to center median bulb | 49.3 ± 2.8 (44.0–53.0) | 47.5 ± 2.4 (45.0–50.0) | 48.0 ± 2.1 (45.0–51.0) | 49.8 ± 2.1 (47.0–52.0) | 49.0 ± 3.4 (43.0–54.0) |
| MB | 51.2 ± 2.2 (47.8–54.1) | 52.4 ± 1.3 (51.1–53.8) | 51.6 ± 1.5 (48.4–53.4) | 53.2 ± 3.8 (50.5–58.8) | 50.0 ± 2.7 (46.7–53.8) |
| Nerve ring to anterior end | 68.6 ± 6.5 (60.0–80.0) | 65.0 ± 4.4 (60.0–70.0) | 65.1 ± 4.1 (59.0–70.0) | 69.5 ± 4.4 (64.0–74.0) | 68.3 ± 5.3 (61.0–76.0) |
| Excretory pore to anterior end | 82.6 ± 8.1 (69.0–97.0) | 85.5 ± 6.6 (79.0–93.0) | 81.0 ± 6.3 (70.0–92.0) | 84.3 ± 6.7 (75.0–90.0) | 84.0 ± 7.3 (75.0–97.0) |
| Pharynx length | 94.8 ± 7.1 (84.0–108.0) | 90.8 ± 4.9 (85.0–97.0) | 93.0 ± 3.8 (88.0–101.0) | 94.0 ± 9.4 (80.0–100.0) | 95.4 ± 7.0 (81.0–105.0) |
| Maximum body diam. | 18.7 ± 2.7 (15.0–23.0) | 18.6 ± 1.5 (16.5–20.0) | 18.4 ± 1.7 (15.0–20.0) | 17.1 ± 0.6 (16.5–18.0) | 18.5 ± 2.3 (16.5–24.0) |
| Tail length | 25.9 ± 4.6 (18.0–36.5) | 29.6 ± 1.3 (28.5–31.5) | 24.5 ± 4.0 (16.5–30.0) | 27.6 ± 1.1 (26.0–28.5) | 26.1 ± 1.7 (23.0–28.0) |
| Anal body diam. | 10.4 ± 1.4 (8.0–13.0) | 11.3 ± 1.6 (10.0–13.5) | 10.0 ± 0.9 (8.0–11.0) | 10.0 ± 0.4 (9.5–10.5) | 9.4 ± 0.8 (8.0–11.0) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clavero-Camacho, I.; Cantalapiedra-Navarrete, C.; Archidona-Yuste, A.; Castillo, P.; Palomares-Rius, J.E. Remarkable Cryptic Diversity of Paratylenchus spp. (Nematoda: Tylenchulidae) in Spain. Animals 2021, 11, 1161. https://doi.org/10.3390/ani11041161
Clavero-Camacho I, Cantalapiedra-Navarrete C, Archidona-Yuste A, Castillo P, Palomares-Rius JE. Remarkable Cryptic Diversity of Paratylenchus spp. (Nematoda: Tylenchulidae) in Spain. Animals. 2021; 11(4):1161. https://doi.org/10.3390/ani11041161
Chicago/Turabian StyleClavero-Camacho, Ilenia, Carolina Cantalapiedra-Navarrete, Antonio Archidona-Yuste, Pablo Castillo, and Juan Emilio Palomares-Rius. 2021. "Remarkable Cryptic Diversity of Paratylenchus spp. (Nematoda: Tylenchulidae) in Spain" Animals 11, no. 4: 1161. https://doi.org/10.3390/ani11041161






