High-Altitude Stress Orchestrates mRNA Expression and Alternative Splicing of Ovarian Follicle Development Genes in Tibetan Sheep
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal and Tissue Samples
2.2. Library Construction and Sequencing
2.3. Analysis of RNA-Seq
2.4. Analysis of Alternative Splicing
2.5. Experimental Validation
3. Results
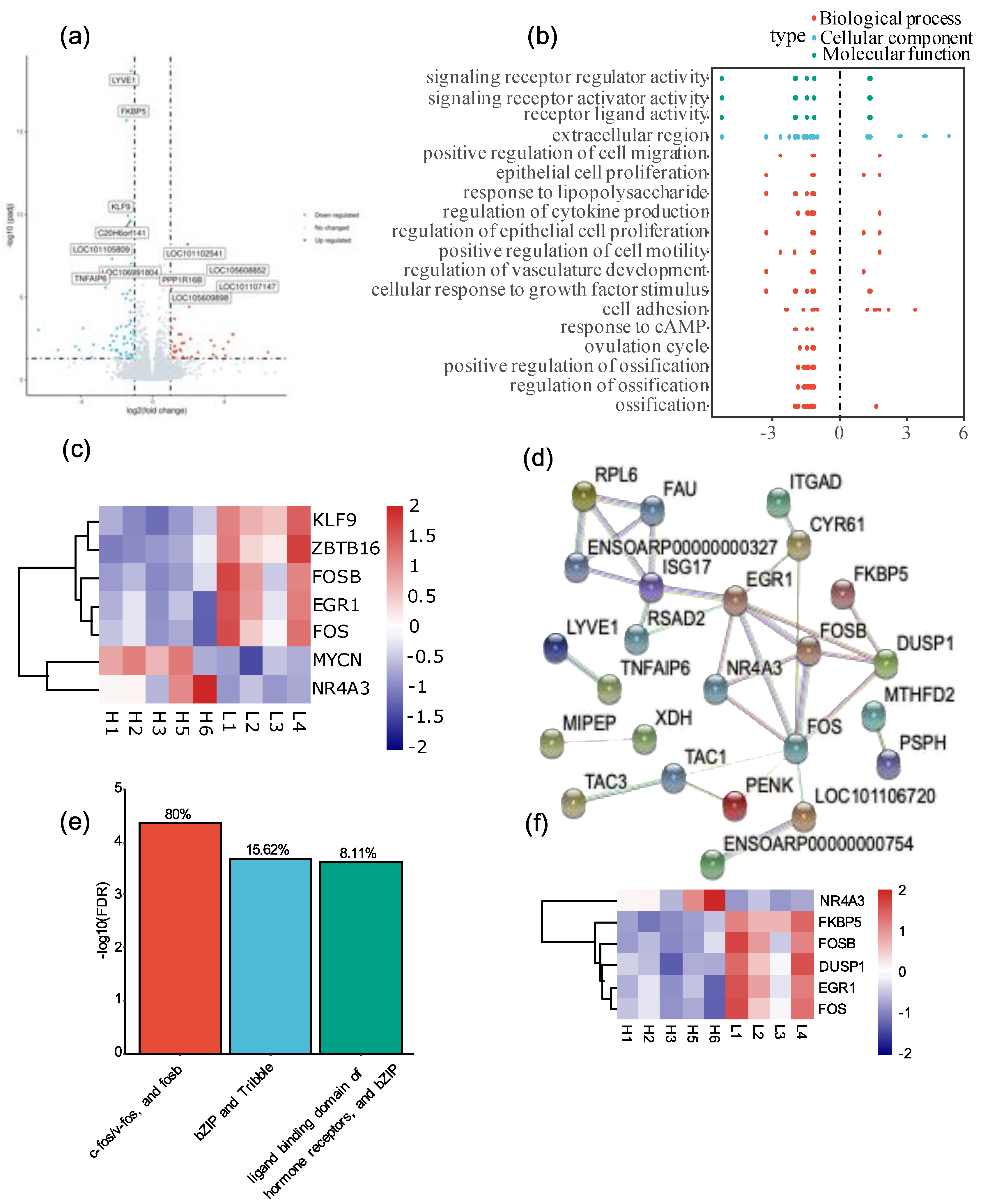
3.1. High-Altitude Stress Affected Gene Expression in Tibetan Sheep
3.2. High-Altitude Stress Repressed the Expression of Follicle Development Genes
3.3. Hormone Signaling Transcription Factors Were Central Regulators in Response to High-Altitude Stress
3.4. Differential Expressed Genes Were Validated by qRT-PCR Experiments
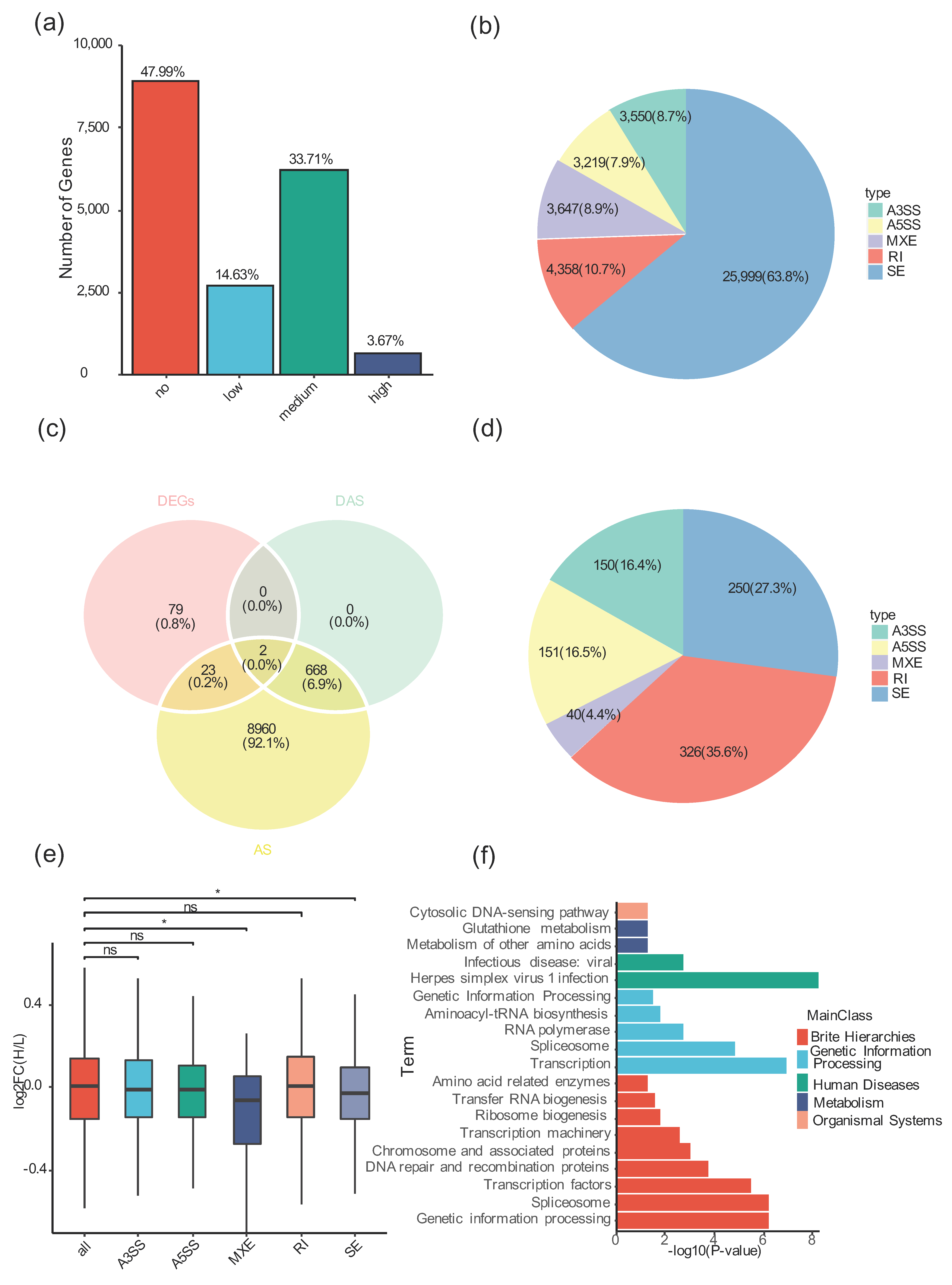
3.5. Alternative Splicing Is Widespread in Tibetan Sheep
3.6. High-Altitude Stress Induced Differential Alternative Splicing in an AS-Type-Specific Pattern
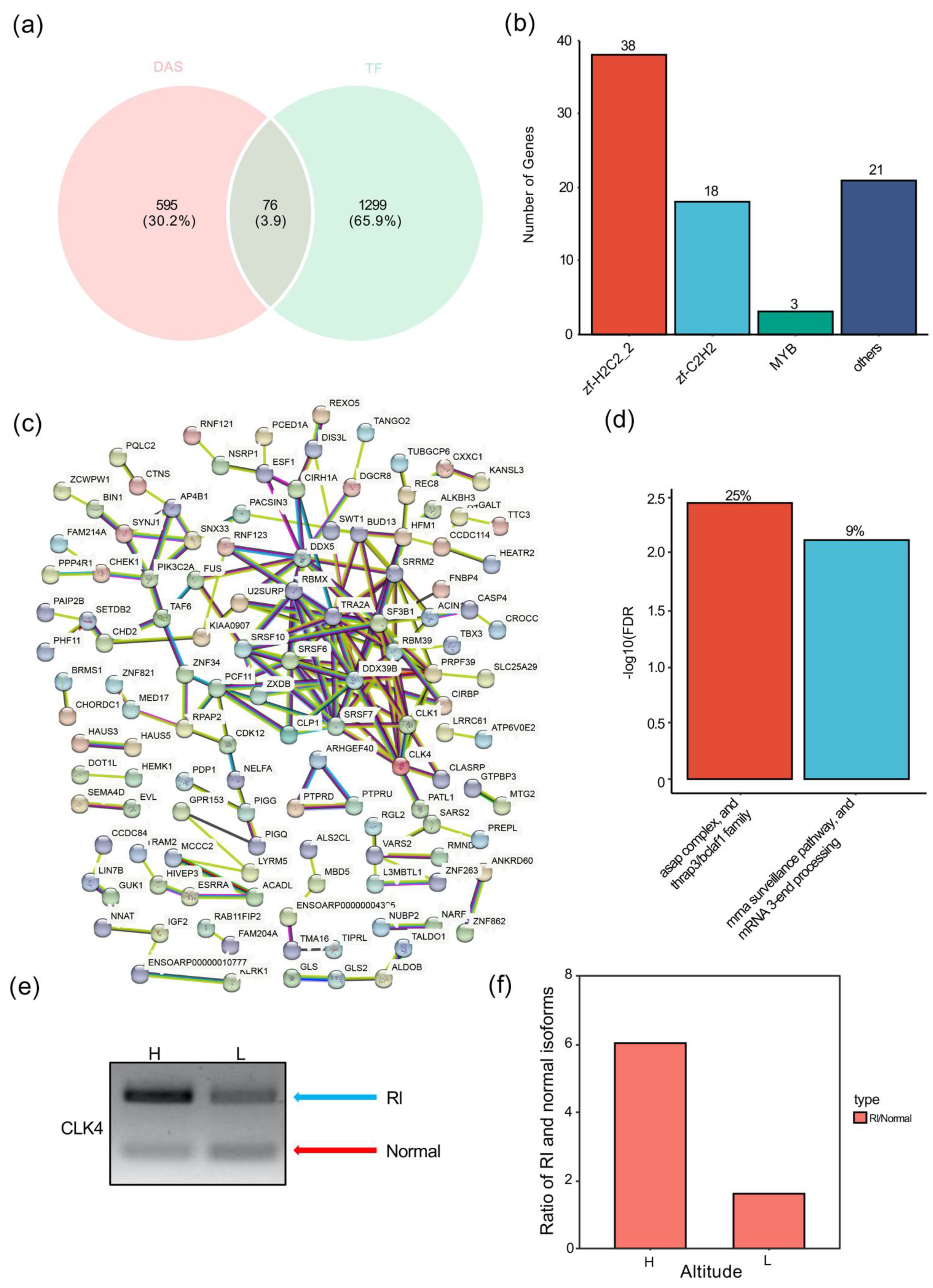
3.7. C2H2-Type Zinc Finger Transcription Factors Enriched in High-Altitude Stress-Related DAS
3.8. High-Altitude Stress-Associated Retained Introns Were Enriched in RNA Processing Factors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Parraguez, V.H.; Gonzalez-Bulnes, A. Endocrinology of reproductive function and pregnancy at high altitudes. Curr. Opin. Endocr. Metabol. Res. 2020, 11, 27–32. [Google Scholar] [CrossRef]
- Parraguez, V.H.; Urquieta, B.; Pérez, L.; Castellaro, G.; De los Reyes, M.; Torres-Rovira, L.; Aguado-Martínez, A.; Astiz, S.; González-Bulnes, A. Fertility in a high-altitude environment is compromised by luteal dysfunction: The relative roles of hypoxia and oxidative stress. Reprod. Biol. Endocrinol. 2013, 11, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Xue, X.; Liu, Y.; Abied, A.; Ding, Y.; Zhao, S.; Wang, W.; Ma, L.; Guo, J.; Guan, W.; et al. Genome-wide comparative analyses reveal selection signatures underlying adaptation and production in Tibetan and Poll Dorset sheep. Sci. Rep. 2021, 11, 2466. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Sun, H. The total output value of Qinghai Yak, Tibetan Sheep Youte Industrial Cluster reached 26.7 billion. China Animal Husbandry and Veterinary News, 3 April 2022; 001. [Google Scholar] [CrossRef]
- Zhao, Y.-X.; Yang, J.; Lv, F.-H.; Hu, X.-J.; Xie, X.-L.; Zhang, M.; Li, W.-R.; Liu, M.-J.; Wang, Y.-T.; Li, J.-Q.; et al. Genomic Reconstruction of the History of Native Sheep Reveals the Peopling Patterns of Nomads and the Expansion of Early Pastoralism in East Asia. Mol. Biol. Evolut. 2017, 34, 2380–2395. [Google Scholar] [CrossRef] [Green Version]
- Hua, Z. Research on efficient breeding model in Tibetan sheep. Chin. Qinghai J. Anim. Vet. Sci. 2016, 46, 28–30. [Google Scholar]
- Wang, H.; Li, X.; Zhou, R.; Xi, J.; Wei, Y.; Li, L.; Zhang, Z. Genome-wide transcriptome profiling in ovaries of small-tail Han sheep during the follicular and luteal phases of the oestrous cycle. Anim. Reprod. Sci. 2018, 197, 212–221. [Google Scholar] [CrossRef]
- Haire, A.; Bai, J.; Zhao, X.; Song, Y.; Zhao, G.; Dilixiati, A.; Li, J.; Sun, W.Q.; Wan, P.; Fu, X.; et al. Identifying the heat resistant genes by multi-tissue transcriptome sequencing analysis in Turpan Black sheep. Theriogenology 2022, 179, 78–86. [Google Scholar] [CrossRef]
- Pokharel, K.; Peippo, J.; Honkatukia, M.; Seppälä, A.; Rautiainen, J.; Ghanem, N.; Hamama, T.-M.; Crowe, M.A.; Andersson, M.; Li, M.-H.; et al. Integrated ovarian mRNA and miRNA transcriptome profiling characterizes the genetic basis of prolificacy traits in sheep (Ovis aries). BMC Genom. 2018, 19, 104. [Google Scholar] [CrossRef] [Green Version]
- Baralle, F.E.; Giudice, J. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 2017, 18, 437–451. [Google Scholar] [CrossRef]
- Yao, S.; Yuan, C.; Shi, Y.; Qi, Y.; Sridha, R.; Dai, M.; Cai, H. Alternative Splicing: A New Therapeutic Target for Ovarian Cancer. Technol. Cancer Res. Treat. 2022, 21, 15330338211067911. [Google Scholar] [CrossRef]
- Ye, W.; Wang, T.; Wei, W.; Lou, S.; Lan, F.; Zhu, S.; Li, Q.; Ji, G.; Lin, C.; Wu, X.; et al. The Full-Length Transcriptome of Spartina alterniflora Reveals the Complexity of High Salt Tolerance in Monocotyledonous Halophyte. Plant Cell Physiol. 2020, 61, 882–896. [Google Scholar] [CrossRef] [PubMed]
- Li, B.J.; Zhu, Z.X.; Qin, H.; Meng, Z.N.; Lin, H.R.; Xia, J.H. Genome-Wide Characterization of Alternative Splicing Events and Their Responses to Cold Stress in Tilapia. Front. Genet. 2020, 11, 244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Yang, X.; Liu, C.; Li, X.; Zhang, B.; Wang, B.; Zhang, Y.; Song, C.; Zhang, T.; Liu, M.; et al. Acetylation of PHF5A Modulates Stress Responses and Colorectal Carcinogenesis through Alternative Splicing-Mediated Upregulation of KDM3A. Mol. Cell 2019, 74, 1250–1263.e6. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Dong, X.; Cao, Z.; Qiu, S.; Li, Y.; Zhong, M.; Xue, Z.; Xu, Y.; Xing, H.; Tang, K.; et al. Mutant U2AF1-induced differential alternative splicing causes an oxidative stress in bone marrow stromal cells. Exp. Biol. Med. 2021, 246, 1750–1759. [Google Scholar] [CrossRef]
- Lu, L.; Yang, Z. Germplasm Characteristics of Plateau Tibetan Sheep in Tianjun County. Shanghai J. Anim. Husband. Vet. Med. 2012, 03, 70–71. [Google Scholar]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen Lars, J.; et al. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, H.; Miao, Y.-R.; Jia, L.-H.; Yu, Q.-Y.; Zhang, Q.; Guo, A.-Y. AnimalTFDB 3.0: A comprehensive resource for annotation and prediction of animal transcription factors. Nucleic Acids Res. 2019, 47, D33–D38. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Ramírez, F.; Ryan, D.P.; Grüning, B.; Bhardwaj, V.; Kilpert, F.; Richter, A.S.; Heyne, S.; Dündar, F.; Manke, T. deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016, 44, W160–W165. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [Green Version]
- Niknafs, Y.S.; Pandian, B.; Iyer, H.K.; Chinnaiyan, A.M.; Iyer, M.K. TACO produces robust multisample transcriptome assemblies from RNA-seq. Nat. Methods 2017, 14, 68–70. [Google Scholar] [CrossRef] [Green Version]
- Pertea, G.; Pertea, M. GFF Utilities: GffRead and GffCompare. F1000Research 2020, 9, 304. [Google Scholar] [CrossRef]
- Shen, S.; Park Juw, W.; Lu, Z.-X.; Lin, L.; Henry Michael, D.; Wu Ying, N.; Zhou, Q.; Xing, Y. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Nat. Acad. Sci. USA 2014, 111, E5593–E5601. [Google Scholar] [CrossRef] [Green Version]
- Lin, Z.; Guo, C.; Lou, S.; Jin, S.; Zeng, W.; Guo, Y.; Fang, J.; Xu, Z.; Zuo, Z.; Ma, L. Functional analyses unveil the involvement of moso bamboo (Phyllostachys edulis) group I and II NIN-LIKE PROTEINS in nitrate signaling regulation. Plant Sci. 2021, 306, 110862. [Google Scholar] [CrossRef]
- Brown, H.M.; Robker, R.L.; Russell, D.L. Development and Hormonal Regulation of the Ovarian Lymphatic Vasculature. Endocrinology 2010, 151, 5446–5455. [Google Scholar] [CrossRef] [PubMed]
- Brown, H.M.; Russell, D.L. Blood and lymphatic vasculature in the ovary: Development, function and disease. Hum. Reprod. Update 2014, 20, 29–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petersen, T.S.; Stahlhut, M.; Andersen, C.Y. Phosphodiesterases in the rat ovary: Effect of cAMP in primordial follicles. Reproduction 2015, 150, 11–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menon, K.M.; Menon, B. Structure, function and regulation of gonadotropin receptors—A perspective. Mol. Cell. Endocrinol. 2012, 356, 88–97. [Google Scholar] [CrossRef] [Green Version]
- Field, S.L.; Dasgupta, T.; Cummings, M.; Orsi, N.M. Cytokines in ovarian folliculogenesis, oocyte maturation and luteinisation. Mol. Reprod. Dev. 2014, 81, 284–314. [Google Scholar] [CrossRef]
- Magata, F.; Horiuchi, M.; Echizenya, R.; Miura, R.; Chiba, S.; Matsui, M.; Miyamoto, A.; Kobayashi, Y.; Shimizu, T. Lipopolysaccharide in ovarian follicular fluid influences the steroid production in large follicles of dairy cows. Anim. Reprod. Sci. 2014, 144, 6–13. [Google Scholar] [CrossRef]
- Choi, Y.; Rosewell, K.L.; Brännström, M.; Akin, J.W.; Curry, T.E., Jr.; Jo, M. FOS, a Critical Downstream Mediator of PGR and EGF Signaling Necessary for Ovulatory Prostaglandins in the Human Ovary. J. Clin. Endocrinol. Metab. 2018, 103, 4241–4252. [Google Scholar] [CrossRef] [Green Version]
- Jackson, D.G.; Prevo, R.; Clasper, S.; Banerji, S. LYVE-1, the lymphatic system and tumor lymphangiogenesis. Trends Immunol. 2001, 22, 317–321. [Google Scholar] [CrossRef]
- Tsubota, K.; Kanki, M.; Noto, T.; Nakatsuji, S.; Oishi, Y.; Matsumoto, M.; Nakayama, H. Altered gene expression profile in ovarian follicle in rats treated with indomethacin and RU486. J. Toxicol. Sci. 2015, 40, 413–425. [Google Scholar] [CrossRef] [Green Version]
- Brown, H.M.; Dunning, K.R.; Robker, R.L.; Pritchard, M.; Russell, D.L. Requirement for ADAMTS-1 in extracellular matrix remodeling during ovarian folliculogenesis and lymphangiogenesis. Dev. Biol. 2006, 300, 699–709. [Google Scholar] [CrossRef]
- Majmundar, A.J.; Wong, W.J.; Simon, M.C. Hypoxia-Inducible Factors and the Response to Hypoxic Stress. Mol. Cell 2010, 40, 294–309. [Google Scholar] [CrossRef] [PubMed]
- Han, P.; Guerrero-Netro, H.; Estienne, A.; Cao, B.; Price, C.A. Regulation and action of early growth response 1 in bovine granulosa cells. Reproduction 2017, 154, 547–557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sayasith, K.; Bouchard, N.; Doré, M.; Sirois, J. Regulation of Bovine Tumor Necrosis Factor-α-Induced Protein 6 in Ovarian Follicles during the Ovulatory Process and Promoter Activation in Granulosa Cells. Endocrinology 2008, 149, 6213–6225. [Google Scholar] [CrossRef] [Green Version]
- Kang, E.; Kim, K.; Jeon, S.Y.; Jung, J.G.; Kim, H.-K.; Lee, H.-B.; Han, W. Targeting CLK4 inhibits the metastasis and progression of breast cancer by inactivating TGF-β pathway. Cancer Gene Ther. 2022, 29, 168–1180. [Google Scholar] [CrossRef]
- Park, M.; Park, S.H.; Park, H.; Kim, H.-R.; Lim, H.J.; Song, H. ADAMTS-1: A novel target gene of an estrogen-induced transcription factor, EGR1, critical for embryo implantation in the mouse uterus. Cell Biosci. 2021, 11, 155. [Google Scholar] [CrossRef] [PubMed]
- Veraguas, D.; Gallegos, P.F.; Velasquez, A.E.; Castro, F.O.; Rodriguez-Alvarez, L. FSH stimulation of anestrous cats improves oocyte quality and development of parthenogenetic embryos. Theriogenology 2017, 87, 25–35. [Google Scholar] [CrossRef]
- Nagyova, E.; Nemcova, L.; Prochazka, R. Expression of tumor necrosis factor alpha-induced protein 6 messenger RNA in porcine preovulatory ovarian follicles. J. Reprod Dev. 2009, 55, 231–235. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Li, Y.; Qiu, C.; Chen, J.; Wu, H.; Wang, Q.; Ma, X.; Song, K.; Kong, B. Splicing Factor DDX23, Transcriptionally Activated by E2F1, Promotes Ovarian Cancer Progression by Regulating FOXM1. Front. Oncol. 2021, 11, 749144. [Google Scholar] [CrossRef]
- Tyzack, G.E.; Neeves, J.; Crerar, H.; Klein, P.; Ziff, O.; Taha, D.M.; Luisier, R.; Luscombe, N.M.; Patani, R. Aberrant cytoplasmic intron retention is a blueprint for RNA binding protein mislocalization in VCP-related amyotrophic lateral sclerosis. Brain 2021, 144, 1985–1993. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Zeng, W.; Jin, X.; Xu, H.; Fang, X.; Ma, Z.; Cao, G.; Li, R.; Ma, L. High-Altitude Stress Orchestrates mRNA Expression and Alternative Splicing of Ovarian Follicle Development Genes in Tibetan Sheep. Animals 2022, 12, 2812. https://doi.org/10.3390/ani12202812
Li W, Zeng W, Jin X, Xu H, Fang X, Ma Z, Cao G, Li R, Ma L. High-Altitude Stress Orchestrates mRNA Expression and Alternative Splicing of Ovarian Follicle Development Genes in Tibetan Sheep. Animals. 2022; 12(20):2812. https://doi.org/10.3390/ani12202812
Chicago/Turabian StyleLi, Wenhao, Weike Zeng, Xiayang Jin, Huiming Xu, Xingyan Fang, Zhijie Ma, Gangjian Cao, Ruizhe Li, and Liuyin Ma. 2022. "High-Altitude Stress Orchestrates mRNA Expression and Alternative Splicing of Ovarian Follicle Development Genes in Tibetan Sheep" Animals 12, no. 20: 2812. https://doi.org/10.3390/ani12202812




