Identification of Potential miRNA–mRNA Regulatory Network Associated with Pig Growth Performance in the Pituitaries of Bama Minipigs and Landrace Pigs
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Source
2.2. Differential Expression Analysis and Enrichment Analysis of DEGs
2.3. Protein–Protein Interaction (PPI) Network Analysis
2.4. miRNA–mRNA Interaction Prediction
2.5. Animal and Cell Culture
2.6. miRNA Transfection
2.7. Quantitative Real-Time PCR (qRT-PCR)
2.8. Statistics Analysis
3. Results
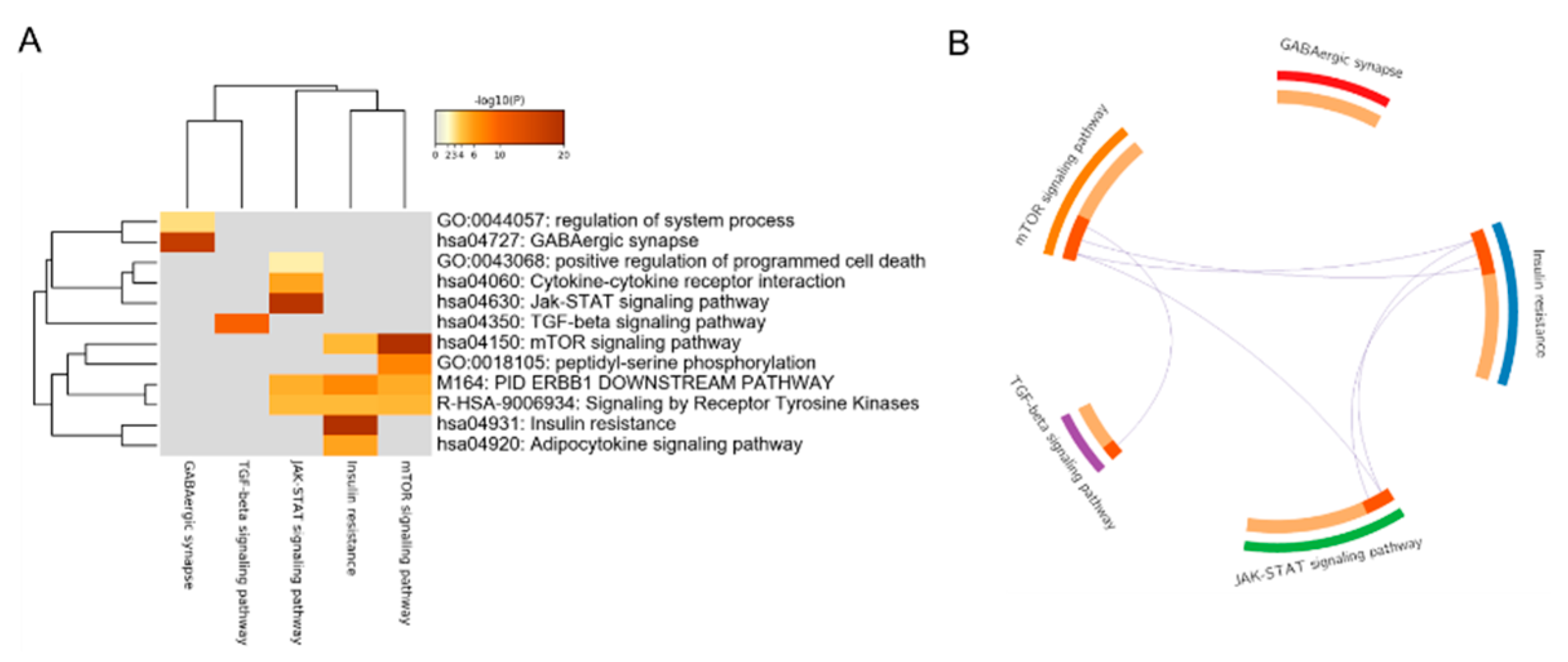
3.1. Gene Set Enrichment Analysis of DEGs
3.2. Construction of the Core PPI Regulation Network
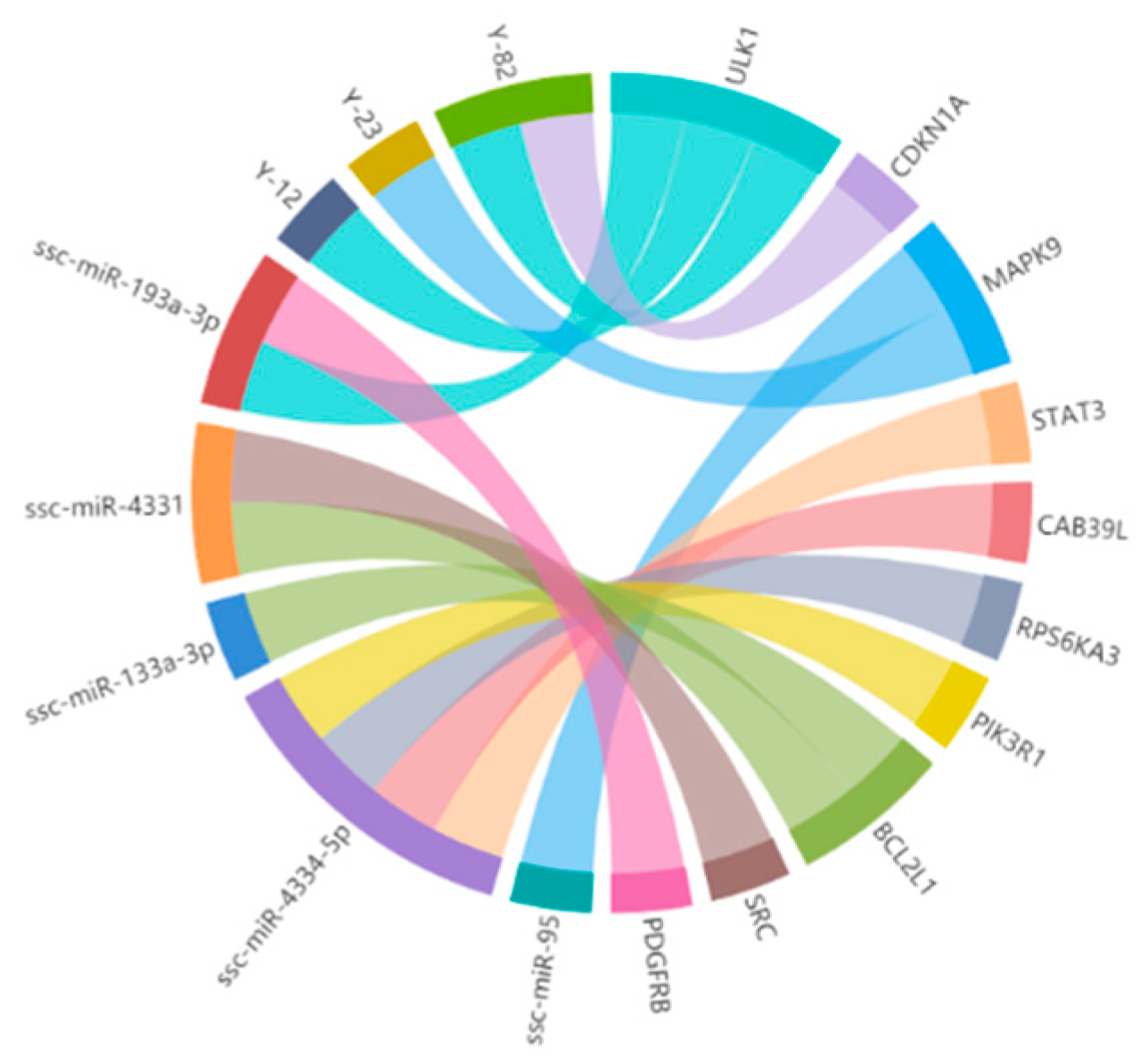
3.3. Construction of the miRNA–mRNA Interaction Network
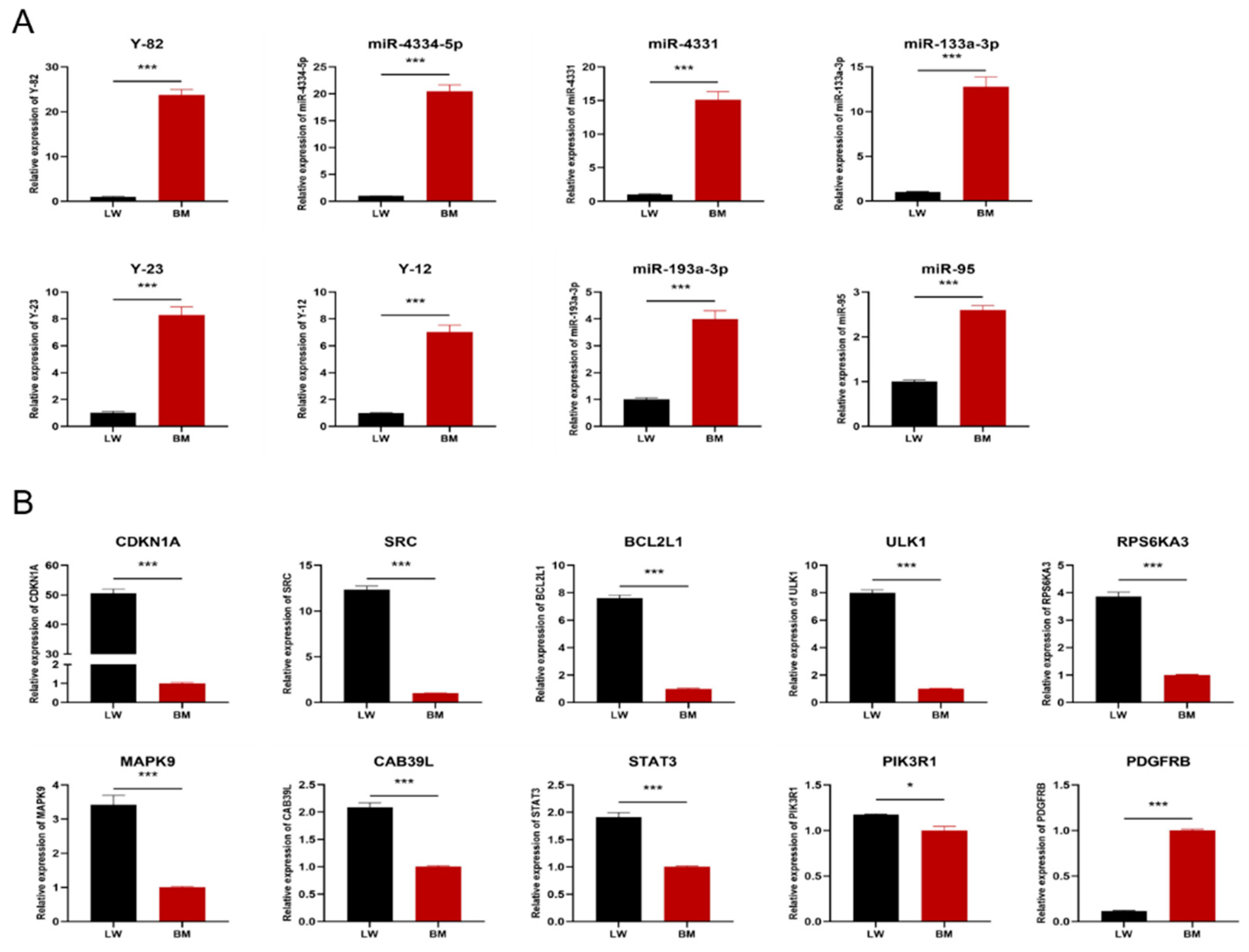
3.4. Validation of Potential Interference Effects of Candidate miRNAs by qRT-PCR
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tejeda, J.F.; Hernandez-Matamoros, A.; Paniagua, M.; Gonzalez, E. Effect of Free-Range and Low-Protein Concentrated Diets on Growth Performance, Carcass Traits, and Meat Composition of Iberian Pig. Animals 2020, 10, 273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, Z.; Zhong, Y.; Yin, Y.; Duan, Y.; Wang, W.; Zhang, L.; Guo, Q.; Chen, Q.; Li, F. Effects of Dietary Tea Powder on the Growth Performance, Carcass Traits, and Meat Quality of Tibetan Pig x Bama Miniature Pigs. Animals 2021, 11, 3225. [Google Scholar] [CrossRef] [PubMed]
- Bonneau, M.; Lebret, B. Production systems and influence on eating quality of pork. Meat Sci. 2010, 84, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Thakali, K.; Morse, P.; Shelby, S.; Chen, J.; Apple, J.; Huang, Y. Comparison of Growth Performance and Meat Quality Traits of Commercial Cross-Bred Pigs versus the Large Black Pig Breed. Animals 2021, 11, 200. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; He, Q.; Azad, M.A.K.; Xiao, Y.; Yin, Y.; Kong, X. Nuclear Magnetic Resonance-Based Metabolomic Analysis Reveals Physiological Stage, Breed, and Diet Effects on the Intramuscular Metabolism of Amino Acids and Related Nutrients in Pigs. Front. Vet. Sci. 2021, 8, 681192. [Google Scholar] [CrossRef] [PubMed]
- Giovannetti, E.; Erozenci, A.; Smit, J.; Danesi, R.; Peters, G.J. Molecular mechanisms underlying the role of microRNAs (miRNAs) in anticancer drug resistance and implications for clinical practice. Crit. Rev. Oncol. Hematol. 2012, 81, 103–122. [Google Scholar] [CrossRef]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef]
- Zhang, H.; Qi, Q.; Chen, T.; Luo, J.; Xi, Q.; Jiang, Q.; Sun, J.; Zhang, Y. Age-Related Changes in MicroRNA in the Rat Pituitary and Potential Role in GH Regulation. Int. J. Mol. Sci. 2018, 19, 2058. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Florez, S.; Gutierrez-Hartmann, A.; Martin, J.F.; Amendt, B.A. MicroRNAs Regulate Pituitary Development, and MicroRNA 26b Specifically Targets Lymphoid Enhancer Factor 1 (Lef-1), Which Modulates Pituitary Transcription Factor 1 (Pit-1) Expression. J. Biol. Chem. 2010, 285, 34718–34728. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Y.; Chen, T.; Song, J.; Qi, Q.; Wang, C.; Xi, Q.; Liu, S.; Hao, L.; Zhang, Y. miR-709 inhibits GHRP6 induced GH synthesis by targeting PRKCA in pituitary. Mol. Cell. Endocrinol. 2020, 506, 110763. [Google Scholar] [CrossRef]
- Ye, R.S.; Li, M.; Qi, Q.E.; Cheng, X.; Chen, T.; Li, C.Y.; Wang, S.B.; Shu, G.; Wang, L.N.; Zhu, X.T.; et al. Comparative Anterior Pituitary miRNA and mRNA Expression Profiles of Bama Minipigs and Landrace Pigs Reveal Potential Molecular Network Involved in Animal Postnatal Growth. PLoS ONE 2015, 10, e0131987. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wei, W.; An, S.; Jiang, J.; He, J.; Zhang, H.; Wang, G.; Han, J.; Liang, B.; Ye, L.; et al. Identification and analysis of lncRNA, microRNA and mRNA expression profiles and construction of ceRNA network in Talaromyces marneffei-infected THP-1 macrophage. PEERJ 2021, 9, e10529. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Furumichi, M.; Morishima, K.; Tanabe, M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019, 47, D590–D595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8, S11. [Google Scholar] [CrossRef] [Green Version]
- Krueger, J.; Rehmsmeier, M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006, 34, W451–W454. [Google Scholar] [CrossRef]
- Liu, Y.; Kong, X.; Jiang, G.; Tan, B.; Deng, J.; Yang, X.; Li, F.; Xiong, X.; Yin, Y. Effects of dietary protein/energy ratio on growth performance, carcass trait, meat quality, and plasma metabolites in pigs of different genotypes. J. Anim. Sci. Biotechnol. 2015, 6, 36. [Google Scholar] [CrossRef] [Green Version]
- Lupu, F.; Terwilliger, J.D.; Lee, K.; Segre, G.V.; Efstratiadis, A. Roles of growth hormone and insulin-like growth factor 1 in mouse postnatal growth. Dev. Biol. 2001, 229, 141–162. [Google Scholar] [CrossRef] [Green Version]
- Eckstrum, K.S.; Weis, K.E.; Baur, N.G.; Yoshihara, Y.; Raetzman, L.T. Icam5 Expression Exhibits Sex Differences in the Neonatal Pituitary and Is Regulated by Estradiol and Bisphenol A. Endocrinology 2016, 157, 1408–1420. [Google Scholar] [CrossRef]
- Lou, W.; Liu, J.; Ding, B.; Chen, D.; Xu, L.; Ding, J.; Jiang, D.; Zhou, L.; Zheng, S.; Fan, W. Identification of potential miRNA-mRNA regulatory network contributing to pathogenesis of HBV-related HCC. J. Transl. Med. 2019, 17, 7. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Lu, F.; Xu, J. Identification of Potential miRNA-mRNA Regulatory Network Contributing to Hypertrophic Cardiomyopathy (HCM). Front. Cardiovasc. Med. 2021, 8, 660372. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Xie, Z.; Zhang, L.; Yang, Y.; Jiang, H.; Ouyang, X.; Zhao, Y.; Liu, Q.; Xu, X.; Li, L. Identification of a Potential miRNA-mRNA Regulatory Network Associated with the Prognosis of HBV-ACLF. Front. Mol. Biosci. 2021, 8, 705683. [Google Scholar] [CrossRef] [PubMed]
- Noroozi, R.; Dinger, M.E.; Fatehi, R.; Taheri, M.; Ghafouri-Fard, S. Identification of miRNA-mRNA Network in Autism Spectrum Disorder Using a Bioinformatics Method. J. Mol. Neurosci. 2021, 71, 761–766. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011, 13, 132U71. [Google Scholar] [CrossRef] [Green Version]
- Kaushik, S.; Rodriguez-Navarro, J.A.; Arias, E.; Kiffin, R.; Sahu, S.; Schwartz, G.J.; Cuervo, A.M.; Singh, R. Autophagy in Hypothalamic AgRP Neurons Regulates Food Intake and Energy Balance. Cell Metab. 2011, 14, 173–183. [Google Scholar] [CrossRef] [Green Version]
- Oh, T.S.; Cho, H.; Cho, J.H.; Yu, S.W.; Kim, E.K. Hypothalamic AMPK-induced autophagy increases food intake by regulating NPY and POMC expression. Autophagy 2016, 12, 2009–2025. [Google Scholar] [CrossRef] [Green Version]
- Song, X.; Zhao, X.; Huang, Y.; Xiang, H.; Zhang, W.; Tong, D. Transmissible Gastroenteritis Virus (TGEV) Infection Alters the Expression of Cellular MicroRNA Species That Affect Transcription of TGEV Gene 7. Int. J. Biol. Sci. 2015, 11, 913–922. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Bai, X.; Guan, L.; Li, J.; Song, X.; Ma, X.; Guo, J.; Zhang, Z.; Du, Q.; Huang, Y.; et al. microRNA-4331 Promotes Transmissible Gastroenteritis Virus (TGEV)-induced Mitochondrial Damage Via Targeting RB1, Upregulating Interleukin-1 Receptor Accessory Protein (IL1RAP) and Activating p38 MAPK Pathway In Vitro. Mol. Cell. Proteom. 2018, 17, 190–204. [Google Scholar] [CrossRef] [Green Version]
- Ruegsegger, G.N.; Creo, A.L.; Cortes, T.M.; Dasari, S.; Nair, K.S. Altered mitochondrial function in insulin-deficient and insulin-resistant states. J. Clin. Investig. 2018, 128, 3671–3681. [Google Scholar] [CrossRef]
- Tian, X.; Ye, M.; Cao, Y.; Wang, C. Losartan Improves Palmitate-Induced Insulin Resistance in 3T3-L1 Adipocytes Through Upregulation of Src Phosphorylation. Exp. Clin. Endocrinol. Diabetes 2017, 125, 136–140. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.T.; Wang, T.Z.; Leng, J.; Chen, Y.; Liu, J.B.; Liu, Y.; Wang, W.J. Palmitate Contributes to Insulin Resistance through Downregulation of the Src-Mediated Phosphorylation of Akt in C2C12 Myotubes. Biosci. Biotechnol. Biochem. 2012, 76, 1356–1361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tu, H.; Xu, C.; Zhang, W.; Liu, Q.; Rondard, P.; Pin, J.P.; Liu, J. GABA(B) Receptor Activation Protects Neurons from Apoptosis via IGF-1 Receptor Transactivation. J. Neurosci. 2010, 30, 749–759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, S.H.; Zhu, L.L.; Zhang, M.; Li, R.K.; Yang, Q.; Yan, J.Y.; Zhang, C.; Yang, J.Y.; Dong, F.Y.; Dai, M.; et al. GABRP regulates chemokine signalling, macrophage recruitment and tumour progression in pancreatic cancer through tuning KCNN4-mediated Ca2+ signalling in a GABA-independent manner. Gut 2019, 68, 1994–2006. [Google Scholar] [CrossRef]
- Athapaththu, A.M.G.K.; Molagoda, I.M.N.; Jayasooriya, R.G.P.T.; Choi, Y.H.; Jeon, Y.J.; Park, J.H.; Lee, B.J.; Kim, G.Y. Gamma-Aminobutyric Acid (GABA) Promotes Growth in Zebrafish Larvae by Inducing IGF-1 Expression via GABA(A) and GABA(B) Receptors. Int. J. Mol. Sci. 2021, 22, 11254. [Google Scholar] [CrossRef]
- Heldsinger, A.; Grabauskas, G.; Song, I.; Owyang, C. Synergistic Interaction between Leptin and Cholecystokinin in the Rat Nodose Ganglia Is Mediated by PI3K and STAT3 Signaling Pathways Implications for Leptin as A Regulator Of Short Term Satiety. J. Biol. Chem. 2011, 286, 11707–11715. [Google Scholar] [CrossRef] [Green Version]
- Ji, F.; Zhang, H.; Wang, Y.; Li, M.; Xu, W.; Kang, Y.; Wang, Z.; Wang, Z.; Cheng, P.; Tong, D.; et al. MicroRNA-133a, downregulated in osteosarcoma, suppresses proliferation and promotes apoptosis by targeting Bcl-xL and Mcl-1. Bone 2013, 56, 220–226. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Liu, X.; Wang, W.; Li, C. miR-133a-3p promotes apoptosis and induces cell cycle arrest by targeting CREB1 in retinoblastoma. Arch. Med. Sci. 2020, 16, 941–956. [Google Scholar] [CrossRef]
- Tian, X.; Li, L.; Fu, G.; Wang, J.; He, Q.; Zhang, C.; Qin, B.; Wang, J. miR-133a-3p regulates the proliferation and apoptosis of intestinal epithelial cells by modulating the expression of TAGLN2. Exp. Ther. Med. 2021, 22, 824. [Google Scholar] [CrossRef]
- Kim, S.; Park, J.W.; Lee, M.G.; Nam, K.H.; Park, J.H.; Oh, H.; Lee, J.; Han, J.; Yi, S.A.; Han, J.W. Reversine promotes browning of white adipocytes by suppressing miR-133a. J. Cell. Physiol. 2019, 234, 3800–3813. [Google Scholar] [CrossRef]
- Wu, Z.; Liu, H.; Sun, W.; Du, Y.; He, W.; Guo, S.; Chen, L.; Zhao, Z.; Wang, P.; Liang, H.; et al. RNF180 mediates STAT3 activity by regulating the expression of RhoC via the proteasomal pathway in gastric cancer cells. Cell Death Dis. 2020, 11, 881. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Li, Y.; Liu, C.; Zhang, L.; Lv, D.; Weng, Y.; Cheng, Z.; Chen, X.; Zhan, J.; Zhang, H. Isonicotinylation is a histone mark induced by the anti-tuberculosis first-line drug isoniazid. Nat. Commun. 2021, 12, 5548. [Google Scholar] [CrossRef] [PubMed]
- Anjum, R.; Roux, P.P.; Ballif, B.A.; Gygi, S.P.; Blenis, J. The tumor suppressor DAP kinase is a target of RSK-mediated survival signaling. Curr. Biol. 2005, 15, 1762–1767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, M.Y.; Hou, L.J.; Sun, J.J.; Zeng, B.; Xi, Q.Y.; Luo, J.Y.; Chen, T.; Zhang, Y.L. Porcine Milk Exosome MiRNAs Attenuate LPS-Induced Apoptosis through Inhibiting TLR4/NF-kappa B and p53 Pathways in Intestinal Epithelial Cells. J. Agric. Food Chem. 2019, 67, 9477–9491. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.-M.; Cao, Q.; Liu, Q. TLR4 regulates vascular smooth muscle cell proliferation in hypertension via modulation of the NLRP3 inflammasome. Am. J. Transl. Res. 2021, 13, 314–325. [Google Scholar]
- Yuan, J.; Wang, K.; Yi, G.; Ma, M.; Dou, T.; Sun, C.; Qu, L.-J.; Shen, M.; Qu, L.; Yang, N. Genome-wide association studies for feed intake and efficiency in two laying periods of chickens. Genet. Sel. Evol. 2015, 47, 82. [Google Scholar] [CrossRef]



| Gene | Sequences (5′–3′) |
|---|---|
| miR-4331 mimics | Sense: UGUGGCUGUGGUGUAGGCCAGC Antisense: UGGCCUACACCACAGCCACAUU |
| miR-133a-3p mimics | Sense: UUGGUCCCCUUCAACCAGCUG Antisense: GCUGGUUGAAGGGGACCAAUU |
| Y-82 mimics | Sense: UAGGGGGCAGGAGCCGGAGCCCUCU Antisense: AGGGCUCCGGCUCCUGCCCCCUAUU |
| NC mimics | Sense: UUCUCCGAACGUGUCACGUTT Antisense: ACGUGACACGUUCGGAGAATT |
| Gene | Sequences (5′–3′) | Product Length (bp) | TM (°C) | Accession Number |
|---|---|---|---|---|
| ULK1 | Sense: CAGTGCCGCTGTGAAGAAGT Antisense: ACCATGACAAAGTCGTCGGT | 246 | 59 | XM_021072523.1 |
| CDKN1A | Sense: ACTTCGACTTCATCACTGAGACC Antisense: ACATGGTCCTCCTGAGACGT | 184 | 59 | XM_013977858.2 |
| MAPK9 | Sense: GACACGGTATTATCGGGCAC Antisense: TCCTCACAGTCGGCTGAAGT | 212 | 59 | XM_005661441.3 |
| STAT3 | Sense: ATCAAGCAGTTCCTTCAGAGC Antisense: TGGACACGCTTGCGAAC | 212 | 59 | NM_001044580.1 |
| CAB39L | Sense: TCTACAATAGTGGGCTGCTGG Antisense: AGCATAATCCCACAGCGTAAG | 220 | 59 | XM_003130971.6 |
| PIK3R1 | Sense: TACTGTAGCCAACAACGGTATGA Antisense: CGTATTTCCCATCTCGGTGA | 227 | 59 | XM_021076847.1 |
| RPS6KA3 | Sense: TCTACTTGGCTGAACTTGCG Antisense: TCTGAGTGTGACCTCGACGA | 228 | 59 | XM_021080348.1 |
| BCL2L1 | Sense: ACTGTGCGTGGAGAGCGTAG Antisense: TCCGACTGAAGAGCGAACC | 251 | 59 | XM_021077300.1 |
| PDGFRB | Sense: GTGGGCTTTCTCCCTGTTGA Antisense: GTAGGCGTCGGAATCCACTT | 240 | 59 | XM_021085013.1 |
| SRC | Sense: CACCTTCCGACTCCATCCAA Antisense: TGTTGAACTGGGTGCGTGAG | 252 | 59 | XM_021077973.1 |
| β-actin | Sense: TGTGCAGGGTATTCATGTGTCCGA Antisense: CAAGGCAAGTTAACAACCCACGGT | 189 | 59 | XM_021086047 |
| miR-95 | Sense: CGCGTTCAACGGGTATTTATTGAGCA | - | 60 | 100316591 |
| miR-4331 | Sense: TATATATACCCTGGAGTGACGGGGG | - | 60 | 100526379 |
| miR-4334-5p | Sense: CAACTGGCCTACAAAGTCCCAGT | - | 60 | 100526405 |
| miR-193a-3p | Sense: TATGTGGCTGTGGTGTAGGCC | - | 60 | 100498771 |
| miR-133a-3p | Sense: TTGGTCCCCTTCAACCAGCTG | - | 60 | 100316580 |
| Y-12 | Sense: TAGGGTGGAGAGATGGATGGATGG | - | 60 | - |
| Y-23 | Sense: CGGAGTAGGAAGGAGGAGGGAAA | - | 60 | - |
| Y-82 | Sense: TATATATAGGGGGCAGGAGCCGG | - | 60 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiao, Y.; Hao, L.; Xia, P.; Cheng, Y.; Song, J.; Chen, X.; Wang, Z.; Ma, Z.; Zheng, S.; Chen, T.; et al. Identification of Potential miRNA–mRNA Regulatory Network Associated with Pig Growth Performance in the Pituitaries of Bama Minipigs and Landrace Pigs. Animals 2022, 12, 3058. https://doi.org/10.3390/ani12213058
Jiao Y, Hao L, Xia P, Cheng Y, Song J, Chen X, Wang Z, Ma Z, Zheng S, Chen T, et al. Identification of Potential miRNA–mRNA Regulatory Network Associated with Pig Growth Performance in the Pituitaries of Bama Minipigs and Landrace Pigs. Animals. 2022; 12(21):3058. https://doi.org/10.3390/ani12213058
Chicago/Turabian StyleJiao, Yingying, Linlin Hao, Peijun Xia, Yunyun Cheng, Jie Song, Xi Chen, Zhaoguo Wang, Ze Ma, Shuo Zheng, Ting Chen, and et al. 2022. "Identification of Potential miRNA–mRNA Regulatory Network Associated with Pig Growth Performance in the Pituitaries of Bama Minipigs and Landrace Pigs" Animals 12, no. 21: 3058. https://doi.org/10.3390/ani12213058
APA StyleJiao, Y., Hao, L., Xia, P., Cheng, Y., Song, J., Chen, X., Wang, Z., Ma, Z., Zheng, S., Chen, T., Zhang, Y., & Yu, H. (2022). Identification of Potential miRNA–mRNA Regulatory Network Associated with Pig Growth Performance in the Pituitaries of Bama Minipigs and Landrace Pigs. Animals, 12(21), 3058. https://doi.org/10.3390/ani12213058






