Intron Regions as Genetic Markers for Population Genetic Investigations of Opisthorchis viverrini sensu lato and Clonorchis sinensis
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Samples
2.2. Primer Design
2.3. PCR Analysis
2.4. DNA Sequencing and Molecular Cloning
2.5. Data Analysis
3. Results
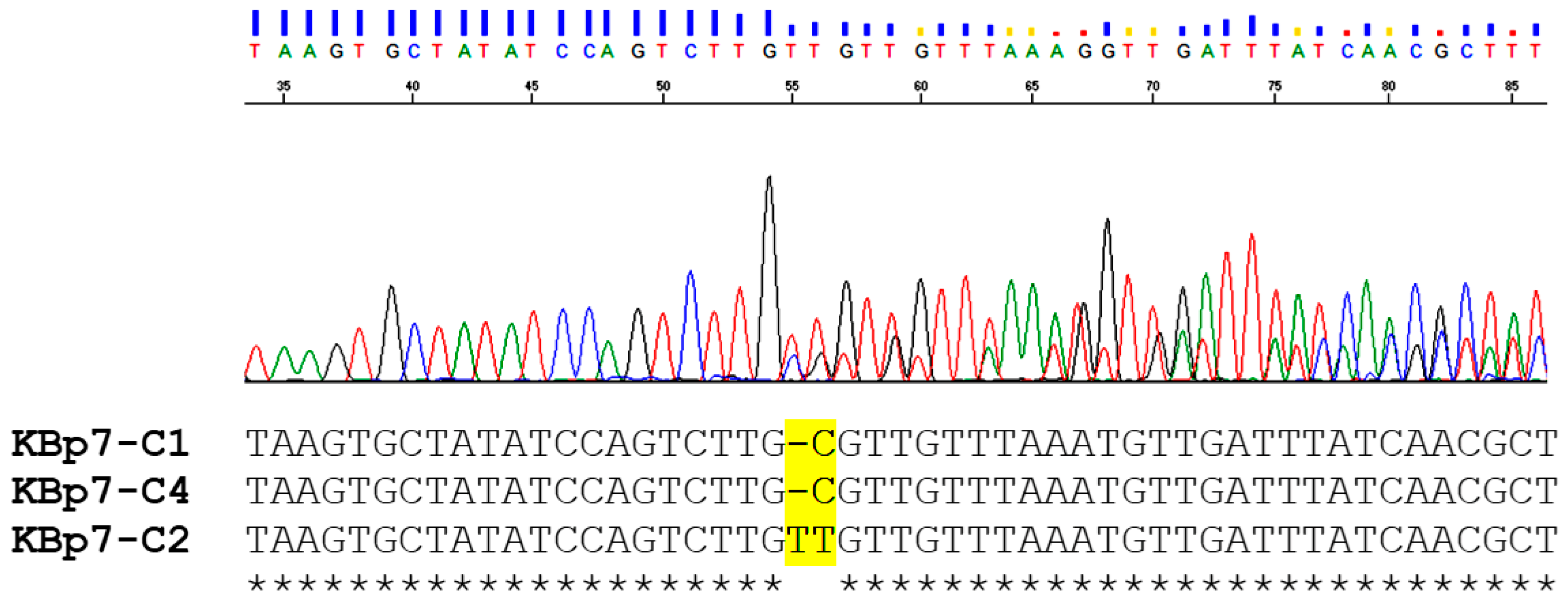
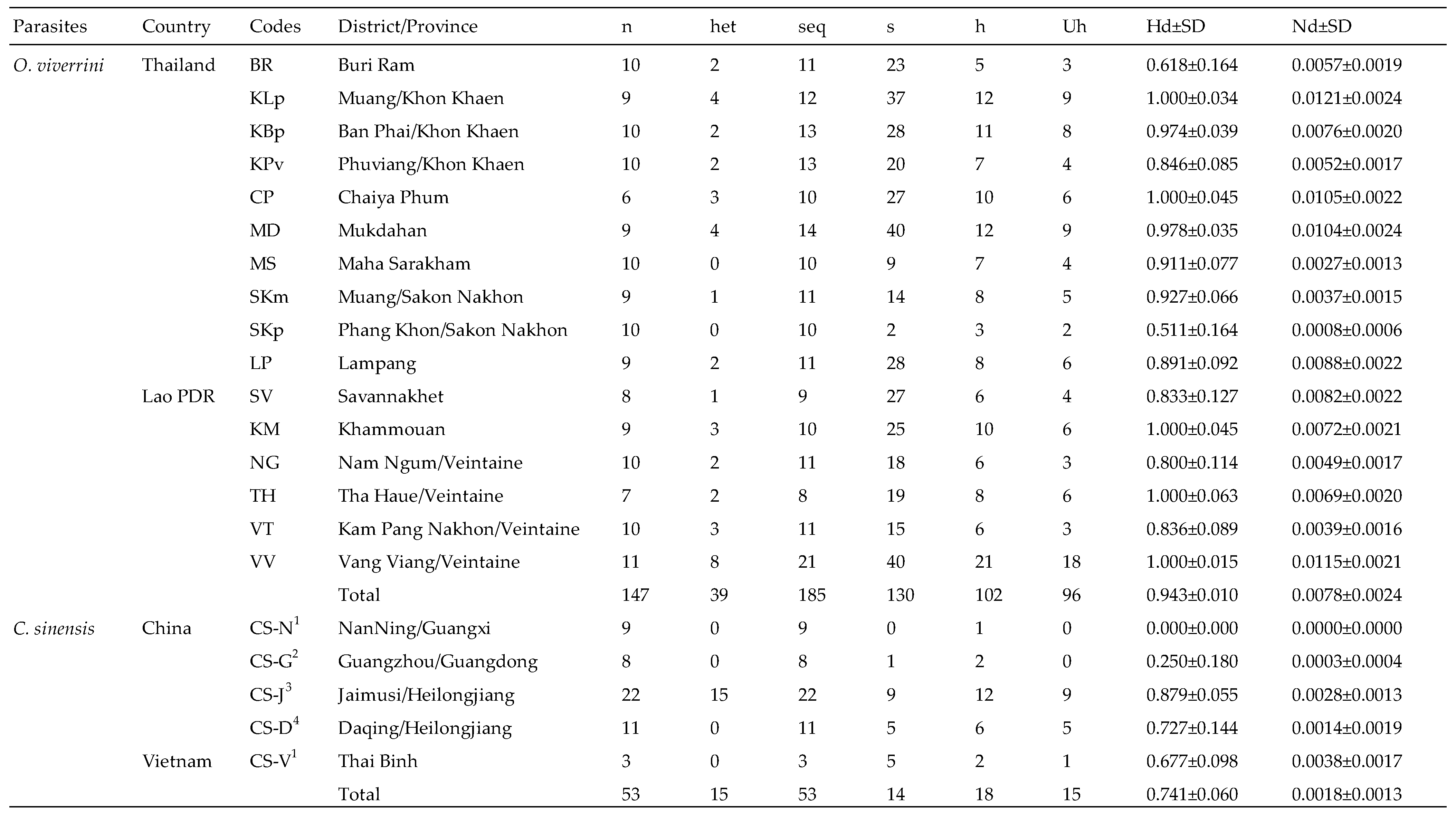
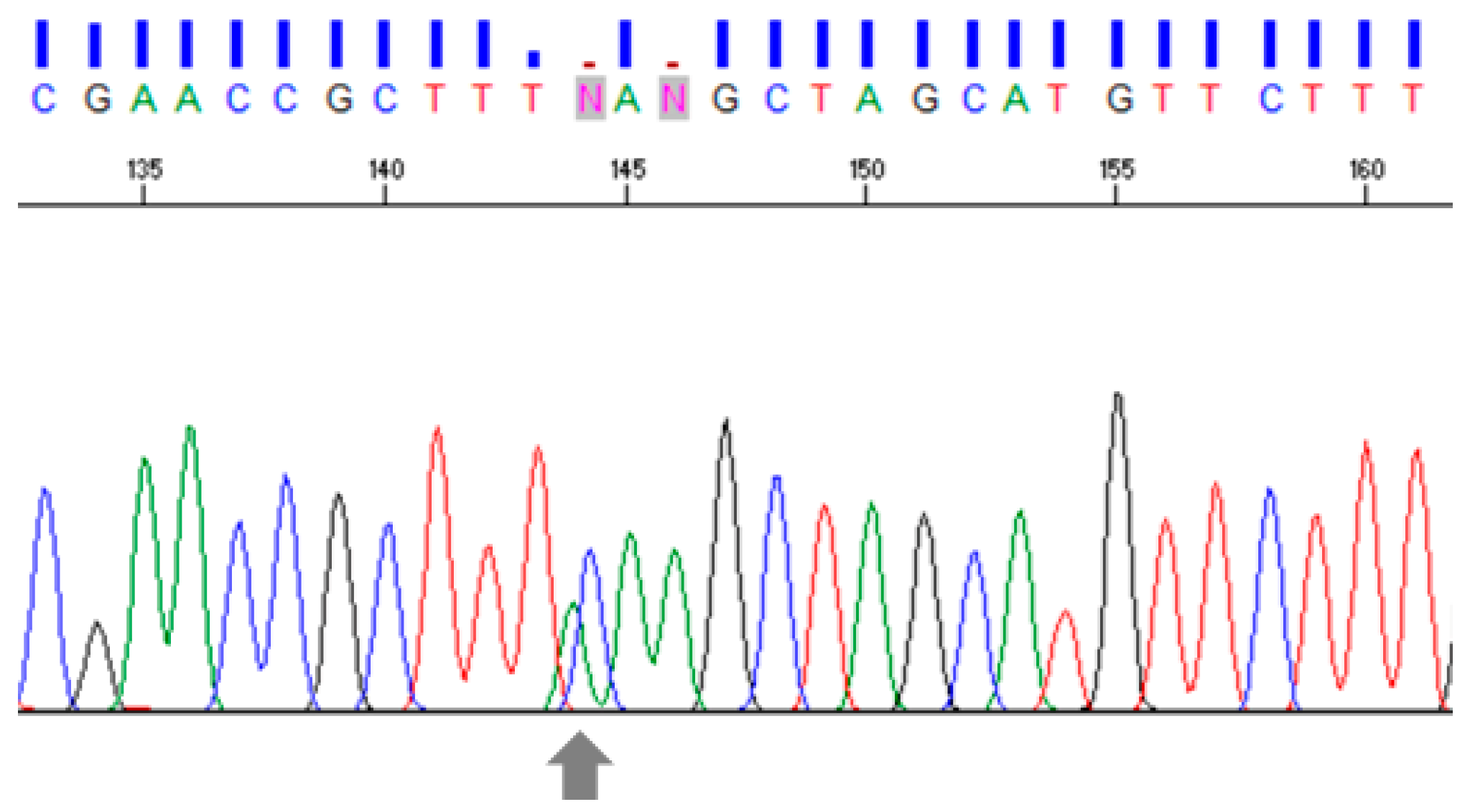
3.1. Intron Amplification
3.2. Nucleotide Sequence Analysis
3.3. Heterozygosity and Phylogenetic Tree Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Control of Foodborne Trematode Infections; World Health Organization: Geneva, Switzerland, 1995. [Google Scholar]
- Keiser, J.; Utzinger, J. Emerging foodborne trematodiasis. Emerg. Infect. Dis. 2005, 11, 1507–1514. [Google Scholar]
- IARC. A review of carcinogen—Part B: Biological Agents. In Monographs on the Evaluation of Carcinogenic Risks to Humans; International Agency for Research on Cancer: Lyon, France, 2011. [Google Scholar]
- Saijuntha, W.; Sithithaworn, P.; Kiatsopit, N.; Andrews, R.H.; Petney, T.N. Liver flukes: Clonorchis and Opisthorchis. In Digenetic Trematodes, 2nd ed.; Toledo, R., Fried, B., Eds.; Advances in Experimental Medicine and Biology; Springer: Cham, Switzerland, 2019; Volume 1154, pp. 139–180. [Google Scholar]
- Sato, P.; Suttiprapa, S.; Tangkawattana, S.; Sripa, M.; Blair, D.; Sripa, B. Does Opisthorchis viverrini circulate between humans and domestic cats in an endemic area in Thailand? Parasitology 2022, 149, 1334–1338. [Google Scholar]
- Wang, D.; Young, N.D.; Korhonen, P.K.; Gasser, R.B. Clonorchis sinensis and Clonorchiasis: The Relevance of Exploring Genetic Variation. Adv. Parasitol. 2018, 100, 155–208. [Google Scholar] [PubMed]
- Saijuntha, W.; Sithithaworn, P.; Petney, T.N.; Andrews, R.H. Current assessment of the systematics and population genetics of Opisthorchis viverrini sensu lato (Trematoda: Opisthorchiidae) and its first intermediate host Bithynia siamensis sensu lato (Gastropoda: Bithyniidae) in Thailand and Southeast Asia. Infect. Genet. Evol. 2022, 97, 105182. [Google Scholar]
- Tang, M.; Zhou, Y.; Liu, Y.; Cheng, N.; Zhang, J.; Xu, X. Molecular identification and genetic-polymorphism analysis of Fasciola flukes in Dali Prefecture, Yunnan Province, China. Parasitol. Int. 2021, 85, 102416. [Google Scholar]
- Platzer, E.G.; Wang, W.; Thompson, S.N.; Borchardt, D.B. Arginine kinase and phosphoarginine, a functional phosphagen, in the rhabditoid nematode steinernema carpocapsae. J. Parasitol. 1999, 85, 603–607. [Google Scholar]
- Uda, K.; Fujimoto, N.; Akiyama, Y.; Mizuta, K.; Tanaka, K.; Ellington, W.R.; Suzuki, T. Evolution of the arginine kinase gene family. Comp. Biochem. Physiol. Part D Genom. Proteom. 2006, 1, 209–218. [Google Scholar]
- Xiao, J.Y.; Lee, J.Y.; Tokuhiro, S.; Ngataki, M.; Jarilla, B.R.; Nomura, H.; Kim, T.I.; Hong, S.J.; Agatsuma, T. Molecular cloning and characterization of taurocyamine kinase from Clonorchis sinensis: A candidate chemotherapeutic target. PLoS Negl. Trop. Dis. 2013, 7, e2548. [Google Scholar]
- Creer, S. Choosing and using introns in molecular phylogenetics. Evol. Bioinform. 2007, 3, 99–108. [Google Scholar]
- Doumen, C. Variable intron/exon structure in the oligochaete lumbricine kinase gene. Gene 2012, 505, 276–282. [Google Scholar]
- Lynch, M. Intron evolution as a population-genetic process. Proc. Natl. Acad. Sci. USA 2002, 99, 6118–6123. [Google Scholar]
- Saijuntha, W.; Tantrawatpan, C.; Wang, C.; Agatsuma, T.; Intapan, P.M.; Maleewong, W.; Petney, T.N. Revealing genetic hybridization and DNA recombination of Fasciola hepatica and Fasciola gigantica in nuclear introns of the hybrid Fasciola flukes. Mol. Biochem. Parasitol. 2018, 223, 31–36. [Google Scholar] [PubMed]
- Tantrawatpan, C.; Tapdara, S.; Agatsuma, T.; Sanpool, O.; Intapan, P.M.; Maleewong, W.; Saijuntha, W. Genetic differentiation of Southeast Asian Paragonimus Braun 1899 (Digenea: Paragonimidae) and genetic variation in the Paragonimus heterotremus complex examined by nuclear DNA sequences. Infect. Genet. Evol. 2021, 90, 104761. [Google Scholar]
- Saijuntha, W.; Tantrawatpan, C.; Agatsuma, T.; Duenngai, K.; Sithithaworn, P.; Andrews, R.H.; Petney, T.N. Intron sequence variation of the echinostomes (Trematoda; Echinostomatidae): Implications for genetic investigations of the 37 collar-spined, Echinostoma miyagawai Ischii, 1932 and E. revolutum (Fröelich, 1802). Parasitol. Res. 2020, 199, 2485–2494. [Google Scholar]
- Prychitko, T.M.; Moore, W.S. Comparative evolution of the mitochondrial cytochrome b gene and nuclear ß-fi brinogen intron 7 in woodpeckers. Mol. Biol. Evol. 2000, 17, 1101–1111. [Google Scholar]
- Prychitko, T.M.; Moore, W.S. Alignment and phylogenetic analysis of β-fi brinogen intron 7 sequences among avian orders reveal conserved regions within the intron. Mol. Biol. Evol. 2003, 20, 762–771. [Google Scholar] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar]
- Kaewkong, W.; Choochote, W.; Kanla, P.; Maleewong, W.; Intapan, P.M.; Wongkham, S.; Wongkham, C. Chromosomes and karyotype analysis of a liver fluke, Opisthorchis viverrini, by scanning electron microscopy. Parasitol. Int. 2012, 61, 504–507. [Google Scholar] [PubMed]
- Thaenkham, U.; Nuamtanong, S.; Vonghachack, Y.; Yoonuan, T.; Sanguankiat, S.; Dekumyoy, P.; Prommasack, B.; Kobayashi, J.; Waikagul, J. Discovery of Opisthorchis lobatus (Trematoda: Opisthorchiidae): A new record of small liver flukes in the Greater Mekong Sub-region. J. Parasitol. 2011, 97, 1152–1158. [Google Scholar] [CrossRef] [PubMed]
- Nawa, Y.; Doanh, P.N.; Thaenkham, U. Is Opisthorchis viverrini an avian liver fluke? J. Helminthol. 2015, 89, 255–256. [Google Scholar] [PubMed]
- Doanh, P.N.; Nawa, Y. Clonorchis sinensis and Opisthorchis spp. in Vietnam: Current status and prospects. Trans. R. Soc. Trop. Med. Hyg. 2016, 110, 13–20. [Google Scholar]
- Dao, T.T.H.; Nguyen, T.T.G.; Gabriel, S.; Bui, K.L.; Dorny, P.; Le, T.H. Updated molecular phylogenetic data for Opisthorchis spp. (Trematoda: Opisthorchioidea) from ducks in Vietnam. Parasit. Vectors 2017, 10, 575. [Google Scholar] [PubMed]
- Chelomina, G.N.; Tatonova, Y.V.; Hung, N.M.; Ngo, H.D. Genetic diversity of the Chinese liver fluke Clonorchis sinensis from Russia and Vietnam. Int. J. Parasitol. 2014, 44, 795–810. [Google Scholar]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tantrawatpan, C.; Maleewong, W.; Thanchomnang, T.; Pilap, W.; Agatsuma, T.; Andrews, R.H.; Sithithaworn, P.; Saijuntha, W. Intron Regions as Genetic Markers for Population Genetic Investigations of Opisthorchis viverrini sensu lato and Clonorchis sinensis. Animals 2023, 13, 3200. https://doi.org/10.3390/ani13203200
Tantrawatpan C, Maleewong W, Thanchomnang T, Pilap W, Agatsuma T, Andrews RH, Sithithaworn P, Saijuntha W. Intron Regions as Genetic Markers for Population Genetic Investigations of Opisthorchis viverrini sensu lato and Clonorchis sinensis. Animals. 2023; 13(20):3200. https://doi.org/10.3390/ani13203200
Chicago/Turabian StyleTantrawatpan, Chairat, Wanchai Maleewong, Tongjit Thanchomnang, Warayutt Pilap, Takeshi Agatsuma, Ross H. Andrews, Paiboon Sithithaworn, and Weerachai Saijuntha. 2023. "Intron Regions as Genetic Markers for Population Genetic Investigations of Opisthorchis viverrini sensu lato and Clonorchis sinensis" Animals 13, no. 20: 3200. https://doi.org/10.3390/ani13203200
APA StyleTantrawatpan, C., Maleewong, W., Thanchomnang, T., Pilap, W., Agatsuma, T., Andrews, R. H., Sithithaworn, P., & Saijuntha, W. (2023). Intron Regions as Genetic Markers for Population Genetic Investigations of Opisthorchis viverrini sensu lato and Clonorchis sinensis. Animals, 13(20), 3200. https://doi.org/10.3390/ani13203200







