Validation of p53 Immunohistochemistry (PAb240 Clone) in Canine Tumors with Next-Generation Sequencing (NGS) Analysis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Case Selection, Histological Classification, and Grading
2.2. Immunohistochemistry
2.3. Next-Generation Sequencing (NGS)
2.4. Statistics
3. Results
3.1. Histopathology and Grading
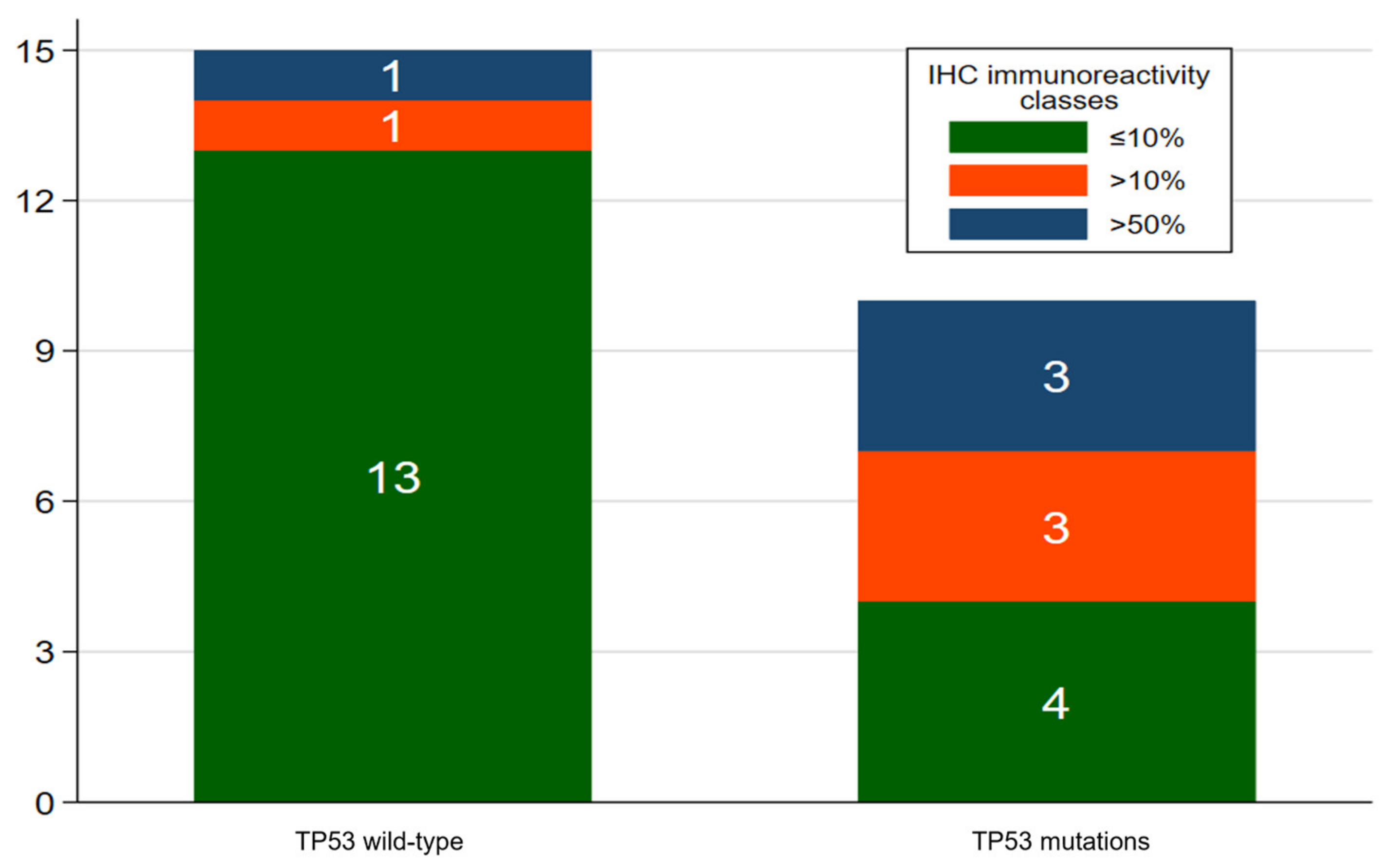
3.2. Immunohistochemistry
3.3. Next-Generation Sequencing (NGS)
3.4. Statistics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hwang, H.J.; Nam, S.K.; Park, H.; Park, Y.; Koh, J.; Na, H.Y.; Kwak, Y.; Kim, W.H.; Lee, H.S. Prediction of TP53 mutations by p53 immunohistochemistry and their prognostic significance in gastric cancer. J. Pathol. Transl. Med. 2020, 54, 378–386. [Google Scholar] [CrossRef] [PubMed]
- Russell, D.S.; Jaworski, L.; Kisseberth, W.C. Immunohistochemical detection of p53, PTEN, Rb, and p16 in canine osteosarcoma using tissue microarray. J. Vet. Diagn. Investig. 2018, 30, 504–509. [Google Scholar] [CrossRef] [PubMed]
- Manoharan, V.; Karunanayake, E.H.; Tennekoon, K.H.; De Silva, S.; Imthikab, A.I.A.; De Silva, K.; Angunawela, P.; Vishwakula, S.; Lunec, J. Pattern of nucleotide variants of TP53 and their correlation with the expression of p53 and its downstream proteins in a Sri Lankan cohort of breast and colorectal cancer patients. BMC Cancer 2020, 20, 72. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Liu, J.; Xu, D.; Zhang, T.; Hu, W.; Feng, Z. Gain-of-function mutant p53 in cancer progression and therapy. J. Mol. Cell Biol. 2020, 12, 674–687. [Google Scholar] [CrossRef]
- Petitjean, A.; Achatz, M.I.W.; Borresen-Dale, A.L.; Hainaut, P.; Olivier, M. TP53 mutations in human cancers: Functional selection and impact on cancer prognosis and outcomes. Oncogene 2007, 26, 2157–2165. [Google Scholar] [CrossRef]
- Lassus, H.; Butzow, R. The classification of p53 immunohistochemical staining results and patient outcome in ovarian cancer. Br. J. Cancer 2007, 96, 1621–1622. [Google Scholar] [CrossRef]
- Köbel, A.M. Interpretation of P53 Immunohistochemistry in Tubo-Ovarian Carcinoma: Guidelines for Reporting; The British Association of Gynaecological Pathologists: London, UK, 2016; pp. 1–17. [Google Scholar]
- Gudkov, A.V.; Komarova, E.A. Pathologies Associated with the p53 Response. Cold Spring Harb. Perspect. Biol. 2010, 2, a001180. [Google Scholar] [CrossRef]
- Suh, Y.; Post, S.M.; Elizondo-Fraire, A.C.; Maccio, D.R.; Jackson, J.G.; El-Naggar, A.K.; Van Pelt, C.; Terzian, T.L. Multiple Stress Signals Activate Mutant P53 in Vivo. Cancer Res. 2011, 1, 7168–7175. [Google Scholar] [CrossRef]
- Köbel, M.; Piskorz, A.M.; Lee, S.; Lui, S.; LePage, C.; Marass, F.; Rosenfeld, N.; Masson, A.-M.M.; Brenton, J.D. Optimized p53 immunohistochemistry is an accurate predictor of TP53 mutation in ovarian carcinoma. J. Pathol. Clin. Res. 2016, 2, 247–258. [Google Scholar] [CrossRef]
- Park, E.; Han, H.; Choi, S.-E.; Park, H.; Woo, H.-Y.; Jang, M.; Shim, H.-S.; Hwang, S.; Kang, H.; Cho, N.-H. p53 Immunohistochemistry and Mutation Types Mismatching in High-Grade Serous Ovarian Cancer. Diagnostics 2022, 12, 579. [Google Scholar] [CrossRef]
- García-Iglesias, M.J.; Cuevas-Higuera, J.L.; Bastida-Sáenz, A.; De Garnica-García, M.G.; Polledo, L.; Perero, P.; González-Fernández, J.; Fernández-Martínez, B.; Pérez-Martínez, C. Immunohistochemical detection of p53 and pp53 Ser392 in canine hemangiomas and hemangiosarcomas located in the skin. BMC Vet. Res. 2020, 16, 239. [Google Scholar] [CrossRef] [PubMed]
- Ilhan, F.; Metin, N.; Birincioglu, S. Immunohistochemical Detection of PCNA and P53 in Mammary Tumours and Normal Tissue in Dogs. Rev. Med. Vet. 2008, 159, 298–304. [Google Scholar]
- Brunetti, B.; Bacci, B.; Angeli, C.; Benazzi, C.; Muscatello, L.V. p53, ER, and Ki67 Expression in Canine Mammary Carcinomas and Correlation with Pathological Variables and Prognosis. Vet. Pathol. 2021, 58, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Munday, J.S.; Ariyarathna, H.; Aberdein, D.; Thomson, N. Immunostaining for p53 and p16CDKN2A Protein Is Not Predictive of Prognosis for Dogs with Malignant Mammary Gland Neoplasms. Vet. Sci. 2019, 6, 34. [Google Scholar] [CrossRef] [PubMed]
- Bongiovanni, L.; Mazzocchetti, F.; Malatesta, D.; Romanucci, M.; Ciccarelli, A.; Buracco, P.; De Maria, R.; Palmieri, C.; Martano, M.; Morello, E.; et al. Immunohistochemical investigation of cell cycle and apoptosis regulators (Survivin, β-Catenin, P53, Caspase 3) in canine appendicular osteosarcoma. BMC Vet. Res. 2012, 8, 78. [Google Scholar] [CrossRef] [PubMed]
- Van Leeuwen, I.; Cornelisse, C.; Misdorp, W.; Goedegebuure, S.; Kirpensteijn, J.; Rutteman, G. P53 gene mutations in osteosarcomas in the dog. Cancer Lett. 1997, 111, 173–178. [Google Scholar] [CrossRef]
- Sagartz, J.E.; Bodley, W.L.; Gamblin, R.M.; Couto, C.G.; Tierney, L.A.; Capen, C.C. p53 Tumor Suppressor Protein Overexpression in Osteogenic Tumors of Dogs. Vet. Pathol. 1996, 33, 213–221. [Google Scholar] [CrossRef]
- Avallone, G.; Muscatello, L.V.; Leoni, A.; Roccabianca, P.; Lepri, E.; Crippa, L.; Bacci, B. p53 Expression in Canine Liposarcoma Correlates with Myxoid Variant and Higher Proliferative Activity. Vet. Pathol. 2020, 57, 620–622. [Google Scholar] [CrossRef]
- Jaffe, M.H.; Hosgood, G.; Taylor, H.W.; Kerwin, S.C.; Hedlund, C.S.; Lopez, M.K.; Davidson, J.R.; Miller, D.M.; Paranjpe, M. Immunohistochemical and Clinical Evaluation of p53 in Canine Cutaneous Mast Cell Tumors. Vet. Pathol. 2000, 37, 40–46. [Google Scholar] [CrossRef]
- Murakami, Y.; Tateyama, S.; Rungsipipat, A.; Uchida, K.; Yamaguchi, R. Immunohistochemical Analysis of Cyclin A, Cyclin D1 and P53 in Mammary Tumors, Squamous Cell Carcinomas and Basal Cell Tumors of Dogs and Cats. J. Vet. Med. Sci. 2000, 62, 743–750. [Google Scholar] [CrossRef]
- Moro, J.V.; Tinuccicosta, M.; Silveira, A.C.T.; Gerardi, D.; Alessi, A. Reactivity of p53 protein in canine transmissible venereal tumor. Arq. Bras. Med. Vet. Zootec. 2010, 62, 318–323. [Google Scholar] [CrossRef]
- Rivera-Calderón, L.G.; Fonseca-Alves, C.E.; Kobayashi, P.E.; Carvalho, M.; Drigo, S.A.; de Oliveira Vasconcelos, R.; Laufer-Amorim, R. Alterations in PTEN, MDM2, TP53 and AR protein and gene expression are associated with canine prostate carcinogenesis. Res. Vet. Sci. 2016, 106, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Wolf, J.C.; Ginn, P.E.; Homer, B.; Fox, L.E.; Kurzman, I.D. Immunohistochemical Detection of p53 Tumor Suppressor Gene Protein in Canine Epithelial Colorectal Tumors. Vet. Pathol. 1997, 34, 394–404. [Google Scholar] [CrossRef]
- Zacchetti, A.; Van Garderen, E.; Rutteman, G.R. Immunohistochemical evaluation of p53 expression with different antibodies in malignant canine tumours with or without p53 gene mutation. Vet. Comp. Oncol. 2007, 5, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Dhaliwal, R.S.; Kitchell, B.E.; Ehrhart, E.; Valli, V.E.; Dervisis, N. Clinicopathologic Significance of Histologic Grade, Pgp, and P53 Expression in Canine Lymphoma. J. Am. Anim. Hosp. Assoc. 2013, 49, 175–184. [Google Scholar] [CrossRef]
- Saito, T.; Chambers, J.K.; Nakashima, K.; Nibe, K.; Ohno, K.; Tsujimoto, H.; Uchida, K.; Nakayama, H. Immunohistochemical analysis of beta-catenin, E-cadherin and p53 in canine gastrointestinal epithelial tumors. J. Vet. Med. Sci. 2020, 82, 1277–1286. [Google Scholar] [CrossRef] [PubMed]
- Zapulli, V.; Peña, L.; Rasotto, R.; Goldschmidt, M.H.; Gama, A.; Scruggs, J.L.; Kiupel, M. Surgical Pathology of Tumors of Domestic Animals Volume 2: Mammary Tumors; Kiupel, M., Ed.; Davis-Thompson DVM Foundation: Gurnee, IL, USA, 2019; ISBN 9781733749114. [Google Scholar]
- Goldschmidt, M.H.; Munday, J.S.; Scruggs, J.L.; Klopfleisch, R.; Kiupel, M. Surgical Pathology of Tumors of Domestic Animals Volume 1: Epithelial Tumors of the Skin; Kiupel, M., Ed.; Davis-Thompson DVM Foundation: Gurnee, IL, USA, 2019. [Google Scholar]
- Roccabianca, P.; Schulman, Y.; Avallone, G.; Foster, R.; Scruggs, J.; Dittmer, K.; Kiupel, M. Surgical Pathology of Tumors of Domestic Animals Volume 3: Tumors of Soft Tissue; Kiupel, M., Ed.; Davis-Thompson DVM Foundation: Gurnee, IL, USA, 2020; ISBN 9781733749121. [Google Scholar]
- Peña, L.; De Andrés, P.J.; Clemente, M.; Cuesta, P.; Pérez-Alenza, M.D. Prognostic Value of Histological Grading in Noninflammatory Canine Mammary Carcinomas in a Prospective Study with Two-Year Follow-Up: Relationship with Clinical and Histological Characteristics. Vet. Pathol. 2013, 50, 94–105. [Google Scholar] [CrossRef]
- Kiupel, M.; Webster, J.D.; Bailey, K.L.; Best, S.; DeLay, J.; Detrisac, C.J.; Fitzgerald, S.D.; Gamble, D.; Ginn, P.E.; Goldschmidt, M.H.; et al. Proposal of a 2-Tier Histologic Grading System for Canine Cutaneous Mast Cell Tumors to More Accurately Predict Biological Behavior. Vet. Pathol. 2011, 48, 147–155. [Google Scholar] [CrossRef]
- Dennis, M.M.; McSporran, K.D.; Bacon, N.J.; Schulman, F.Y.; Foster, R.A.; Powers, B.E. Prognostic Factors for Cutaneous and Subcutaneous Soft Tissue Sarcomas in Dogs. Vet. Pathol. 2010, 48, 73–84. [Google Scholar] [CrossRef]
- Albaric, O.; Bret, L.; Amardeihl, M.; Delverdier, M. Immunohistochemical expression of p53 in animal tumors: A methodological study using four anti-human p53 antibodies. Histol. Histopathol. 2001, 16, 113–121. [Google Scholar]
- Keller, S.; Schade, B.; Rickenbacher, A.; Brugnera, E.; Wergin, M.; Müller, E.; Suter, M.; Guscetti, F. A Comprehensive Test System to Identify Suitable Antibodies Against p53 for Immunohistochemical Analysis of Canine Tissues. J. Comp. Pathol. 2007, 137, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Altman, D.G. Practical Statistics for Medical Research, 2nd ed.; Chapman & Hall/CRC: London, UK, 1991. [Google Scholar]
- Szklo, M.; Nieto, F.J. Epidemiology: Beyond the Basics, 3rd ed.; Jones & Bartlett Learning: Burlington, MA, USA, 2014; ISBN 9781449604707. [Google Scholar]
- Duffy, M.J.; Synnott, N.C.; Crown, J. Mutant p53 as a target for cancer treatment. Eur. J. Cancer 2017, 83, 258–265. [Google Scholar] [CrossRef] [PubMed]
- Perri, F.; Pisconti, S.; Scarpati, G.D.V. P53 mutations and cancer: A tight linkage. Ann. Transl. Med. 2016, 4, 522. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Hayashi, T.; Inoue, M. Immunohistochemical Expression of Mdm2 and p53 in Canine Cutaneous Mast Cell Tumours. J. Vet. Med. Ser. Physiol. Pathol. Clin. Med. 2006, 53, 65–68. [Google Scholar] [CrossRef] [PubMed]
- Ihle, M.A.; Fassunke, J.; König, K.; Grünewald, I.; Schlaak, M.; Kreuzberg, N.; Tietze, L.; Schildhaus, H.; Büttner, R.; Merkelbach-bruse, S. Comparison of high-resolution melting analysis, pyrosequencing, next generation sequencing and immunohistochemistry to conventional Sanger sequencing for the detection of p. V600E and non-p. V600E BRAF mutations. BMC Cancer 2014, 14, 13. [Google Scholar] [CrossRef] [PubMed]
- Roshandel, A.K.; Busch, C.M.; Van Mullekom, J.; Cuoco, J.A.; Rogers, C.M.; Apfel, L.S.; Marvin, E.A.; Sontheimer, H.W.; Umans, R.A. The predictive capability of immunohistochemistry and DNA sequencing for determining TP53 functional mutation status: A comparative study of 41 glioblastoma patients. Oncotarget 2019, 10, 6204–6218. [Google Scholar] [CrossRef]
- Murnyák, B.; Hortobágyi, T. Immunohistochemical correlates of TP53 somatic mutations in cancer. Oncotarget 2016, 7, 64910–64920. [Google Scholar] [CrossRef]
- Nagahashi, M.; Shimada, Y.; Ichikawa, H.; Nakagawa, S.; Sato, N.; Kaneko, K.; Homma, K.; Kawasaki, T.; Kodama, K.; Lyle, S.; et al. Formalin-fixed paraffin-embedded sample conditions for deep next generation sequencing. J. Surg. Res. 2018, 220, 125–132. [Google Scholar] [CrossRef]
- Heydt, C.; Fassunke, J.; Künstlinger, H.; Ihle, M.A.; König, K.; Heukamp, L.; Schildhaus, H.-U.; Odenthal, M.; Büttner, R.; Merkelbach-Bruse, S. Comparison of Pre-Analytical FFPE Sample Preparation Methods and Their Impact on Massively Parallel Sequencing in Routine Diagnostics. PLoS ONE 2014, 9, e104566. [Google Scholar] [CrossRef]

| Tissue | Histotype | Grade | IHC Class | TP53 (Ref. TP53-202) | Mutation | PolyPhen2 Score |
|---|---|---|---|---|---|---|
| Mammary (3) | Comedocarcinoma | 3 | >50% | NE | - | - |
| Solid carcinoma | 2 | 10–50% | NE | - | - | |
| Comedocarcinoma | 3 | >50% | NE | - | - | |
| Skin (6) | SCC | - | 10–50% | p.C290Y | Missense | 1 |
| SCC | - | 10–50% | p.Y257F p.R265Stop p.C329F | Missense Non-sense Missense | 1 1 1 | |
| SCC | - | >50% | p.P202L p.P203S p.R265Stop | Missense Missense Non-sense | 0.969 0.997 1 | |
| SCC | - | >50% | p.P193L p.H230D | Missense Missense | 1 0.993 | |
| SCC | - | 10–50% | WT | - | ||
| SCC | - | 10–50% | NE | - | ||
| Skin (1) | MCT | high | 10–50% | p.R209S | Missense | 1 |
| Oral (2) | Amelanotic melanoma | - | >50% | p.G318E | Missense | 1 |
| Amelanotic melanoma | - | 10–50% | NE | - | ||
| Subcutis (3) | STS | 2 | >50% | WT | - | |
| STS | 2 | >50% | NE | - | ||
| STS | 1 | 10–50% | NE | - |
| Tissue | Histotype | Grade | IHC Class | TP53 (Ref. TP53-202) | Mutation | PolyPhen2 Score |
|---|---|---|---|---|---|---|
| Mammary (2/15) | Solid carcinoma | 3 | <10% | p.C186Y | Missense | 1 |
| Comedocarcinoma | 3 | <10% | p.P372S | Missense | 0.997 | |
| Skin (1/5) | SCC | - | <10% | p.R265Stop | Non-sense | 1 |
| Bone (1/1) | Osteosarcoma | - | <10% | p.R301P | Missense | 1 |
| NGS Mutations | NGS WT | Total | ||
|---|---|---|---|---|
| IHC P53+ | 6 (TP) | 2 (FP) | 8 | PPV 75% (34.9–96.8%) |
| IHC P53- | 4 (FN) | 13 (TN) | 17 | NPV 76.5% (50.1–93.2.4%) |
| Total | 10 | 15 | 25 | |
| Sensitivity 60% (26.2–87.8%) | Specificity 86.7% (59.5–98.3%) | Accuracy 76% (54.9–90.6%) |
| NGS Mutations | NGS WT | Total | ||
|---|---|---|---|---|
| IHC P53+ | 3 (TP) | 1 (FP) | 4 | PPV 75% (19.4–99.4%) |
| IHC P53- | 7 (FN) | 14 (TN) | 21 | NPV 66% (43–85.4%) |
| Total | 10 | 15 | 25 | |
| Sensitivity 30% (6.7–65.2%) | Specificity 93% (68.1–99.8%) | Accuracy 68% (46.5–85.1%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brunetti, B.; de Biase, D.; Dellapina, G.; Muscatello, L.V.; Ingravalle, F.; Tura, G.; Bacci, B. Validation of p53 Immunohistochemistry (PAb240 Clone) in Canine Tumors with Next-Generation Sequencing (NGS) Analysis. Animals 2023, 13, 899. https://doi.org/10.3390/ani13050899
Brunetti B, de Biase D, Dellapina G, Muscatello LV, Ingravalle F, Tura G, Bacci B. Validation of p53 Immunohistochemistry (PAb240 Clone) in Canine Tumors with Next-Generation Sequencing (NGS) Analysis. Animals. 2023; 13(5):899. https://doi.org/10.3390/ani13050899
Chicago/Turabian StyleBrunetti, Barbara, Dario de Biase, Giulia Dellapina, Luisa Vera Muscatello, Francesco Ingravalle, Giorgia Tura, and Barbara Bacci. 2023. "Validation of p53 Immunohistochemistry (PAb240 Clone) in Canine Tumors with Next-Generation Sequencing (NGS) Analysis" Animals 13, no. 5: 899. https://doi.org/10.3390/ani13050899
APA StyleBrunetti, B., de Biase, D., Dellapina, G., Muscatello, L. V., Ingravalle, F., Tura, G., & Bacci, B. (2023). Validation of p53 Immunohistochemistry (PAb240 Clone) in Canine Tumors with Next-Generation Sequencing (NGS) Analysis. Animals, 13(5), 899. https://doi.org/10.3390/ani13050899








