Genetic Variability in Leishmaniasis-Causing Leishmania infantum in Humans and Dogs from North-East Spain
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Human and Dog Samples
2.2. PCR Amplification
2.3. Sequencing and Analysis of Single Nucleotide Polymorphisms
2.4. Restriction Fragment Length Polymorphism Analysis
2.5. Phylogenetic Analysis
3. Results
3.1. Genotypes Obtained by SNP Analysis
3.2. Genotypes Obtained by RFLP Analysis
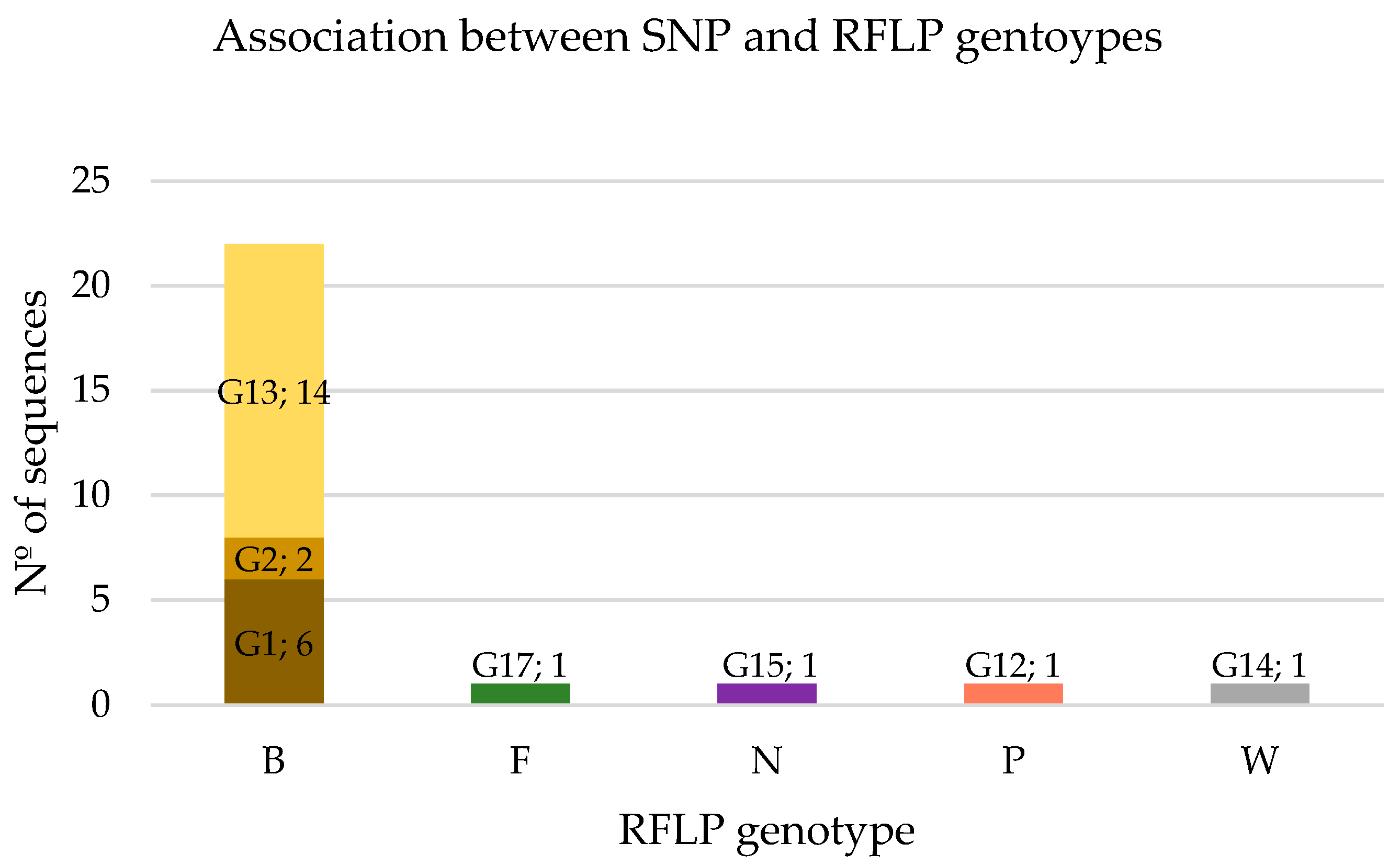
3.3. Association between SNP and RFLP Genotypes
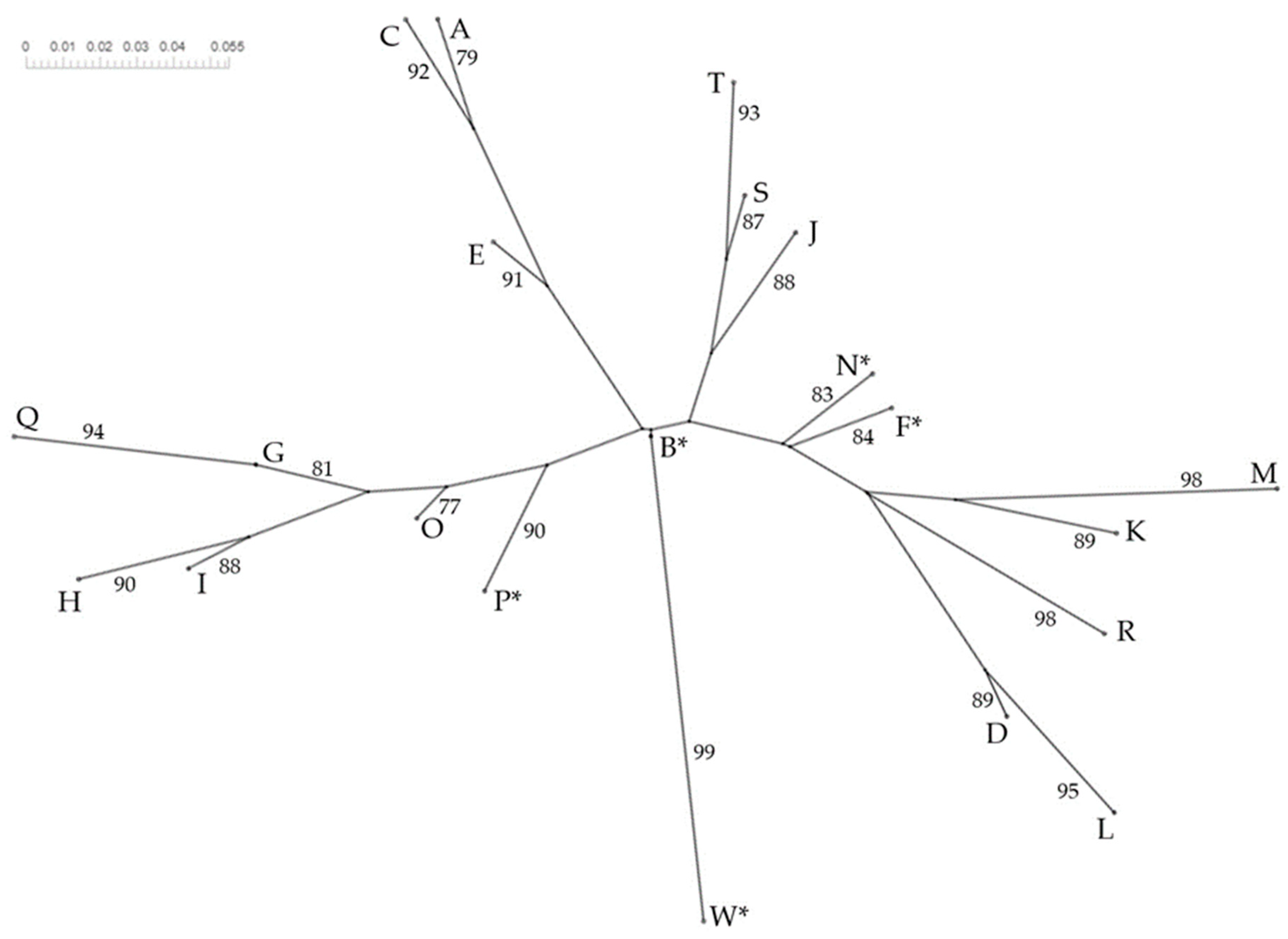
3.4. Phylogenetic Study
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization Leishmaniasis. Available online: https://www.who.int/news-room/fact-sheets/detail/leishmaniasis (accessed on 30 April 2024).
- Cortes, S.; Mauricio, I.; Almeida, A.; Cristovão, J.M.; Pratlong, F.; Dedet, J.P.; Campino, L. Application of KDNA as a Molecular Marker to Analyse Leishmania infantum Diversity in Portugal. Parasitol. Int. 2006, 55, 277–283. [Google Scholar] [CrossRef]
- Alcover, M.M.; Rocamora, V.; Guillén, M.C.; Berenguer, D.; Cuadrado, M.; Riera, C.; Fisa, R. Case Report: Diffuse Cutaneous Leishmaniasis by Leishmania infantum in a Patient Undergoing Immunosuppressive Therapy: Risk Status in an Endemic Mediterranean Area. Am. J. Trop. Med. Hyg. 2018, 98, 1313–1316. [Google Scholar] [CrossRef] [PubMed]
- Alcover, M.M.; Rocamora, V.; Ribas, A.; Fisa, R.; Riera, C. Underestimation of Human Cutaneous Leishmaniasis Caused by Leishmania infantum in an Endemic Area of the Mediterranean Basin (Balearic Islands). Microorganisms 2023, 11, 126. [Google Scholar] [CrossRef]
- Alvar, J.; Aparicio, P.; Aseffa, A.; Den Boer, M.; Cañavate, C.; Dedet, J.P.; Gradoni, L.; Ter Horst, R.; López-Vélez, R.; Moreno, J. The Relationship between Leishmaniasis and AIDS: The Second 10 Years. Clin. Microbiol. Rev. 2008, 21, 334–359. [Google Scholar] [CrossRef]
- Monge-Maillo, B.; Norman, F.F.; Cruz, I.; Alvar, J.; López-Vélez, R. Visceral Leishmaniasis and HIV Coinfection in the Mediterranean Region. PLoS Negl. Trop. Dis. 2014, 8, e3021. [Google Scholar] [CrossRef]
- Alcover, M.M.; Basurco, A.; Fernandez, A.; Riera, C.; Fisa, R.; Gonzalez, A.; Verde, M.; Garrido, A.M.; Ruíz, H.; Yzuel, A.; et al. A Cross-Sectional Study of Leishmania infantum Infection in Stray Cats in the City of Zaragoza (Spain) Using Serology and PCR. Parasit. Vectors 2021, 14, 178. [Google Scholar] [CrossRef] [PubMed]
- Alcover, M.M.; Giner, J.; Rabasedas, J.; Roca-Geronès, X.; Verde, M.; Fernández, A.; Riera, C.; Fisa, R.; Villanueva-Saz, S. First Epidemiological Survey of Leishmania infantum in the Domestic Ferret (Mustela putorius furo) in a Canine Leishmaniosis Endemic Area Using Serology and PCR. Parasit. Vectors 2022, 15, 372. [Google Scholar] [CrossRef] [PubMed]
- Baxarias, M.; Mateu, C.; Miró, G.; Solano-Gallego, L. Serological Survey of Leishmania infantum in Apparently Healthy Dogs in Different Areas of Spain. Vet. Med. Sci. 2023, 9, 1980–1988. [Google Scholar] [CrossRef]
- Díaz-Regañón, D.; Roura, X.; Suárez, M.L.; León, M.; Sainz, Á. Serological Evaluation of Selected Vector-Borne Pathogens in Owned Dogs from Northern Spain Based on a Multicenter Study Using a Commercial Test. Parasit. Vectors 2020, 13, 301. [Google Scholar] [CrossRef]
- Gálvez, R.; Montoya, A.; Cruz, I.; Fernández, C.; Martín, O.; Checa, R.; Chicharro, C.; Migueláñez, S.; Marino, V.; Miró, G. Latest Trends in Leishmania infantum Infection in Dogs in Spain, Part I: Mapped Seroprevalence and Sand Fly Distributions. Parasit. Vectors 2020, 13, 204. [Google Scholar] [CrossRef]
- Miró, G.; Wright, I.; Michael, H.; Burton, W.; Hegarty, E.; Rodón, J.; Buch, J.; Pantchev, N.; von Samson-Himmelstjerna, G. Seropositivity of Main Vector-Borne Pathogens in Dogs across Europe. Parasit. Vectors 2022, 15, 189. [Google Scholar] [CrossRef] [PubMed]
- Montoya-Alonso, J.A.; Morchón, R.; Costa-Rodríguez, N.; Matos, J.I.; Falcón-Cordón, Y.; Carretón, E. Current Distribution of Selected Vector-Borne Diseases in Dogs in Spain. Front. Vet. Sci. 2020, 7, 564429. [Google Scholar] [CrossRef]
- Fernández Martínez, B. Situación de Leishmaniasis En España. Años 2019, 2020 y 2021. Boletín Epidemiológico Sem. 2023, 31, 83–92. [Google Scholar] [CrossRef]
- Departamento de Sanidad del Gobierno de Aragón. Boletín Epidemiológico Semanal de Aragón. Semana 32/2020. Available online: https://www.aragon.es/documents/20127/1650151/BOLETIN+ARAGON+322020.pdf/532edf77-c1c8-94d3-4c22-336e91d98226?t=1597325644731 (accessed on 30 April 2024).
- Schönian, G.; Mauricio, I.; Cupolillo, E. Is It Time to Revise the Nomenclature of Leishmania? Trends Parasitol. 2010, 26, 466–469. [Google Scholar] [CrossRef]
- Van der Auwera, G.; Dujardin, J.C. Species Typing in Dermal Leishmaniasis. Clin. Microbiol. Rev. 2015, 28, 265–294. [Google Scholar] [CrossRef]
- Rioux, J.A.; Lanotte, G.; Serres, E.; Pratlong, F.; Bastien, P.; Perieres, J. Taxonomy of Leishmania. Use of Isoenzymes. Suggestions for a New Classification. Ann. Parasitol. Hum. Comp. 1990, 65, 111–125. [Google Scholar] [CrossRef]
- El Hamouchi, A.; Ejghal, R.; Hida, M.; Lemrani, M. Intraspecific Genetic Variability in a Population of Moroccan Leishmania infantum Revealed by PCR-RFLP of KDNA Minicircles. Acta Trop. 2017, 169, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Marfurt, J.; Nasereddin, A.; Niederwieser, I.; Jaffe, C.L.; Beck, H.P.; Felger, I. Identification and Differentiation of Leishmania Species in Clinical Samples by PCR Amplification of the Miniexon Sequence and Subsequent Restriction Fragment Length Polymorphism Analysis. J. Clin. Microbiol. 2003, 41, 3147–3153. [Google Scholar] [CrossRef]
- Azmi, K.; Nasereddin, A.; Ereqat, S.; Schönian, G.; Abdeen, Z. Identification of Old World Leishmania Species by PCR-RFLP of the 7 Spliced Leader RNA Gene and Reverse Dot Blot Assay. Trop. Med. Int. Health 2010, 15, 872–880. [Google Scholar] [CrossRef]
- Montalvo, A.M.; Fraga, J.; Maes, I.; Dujardin, J.C.; Van Der Auwera, G. Three New Sensitive and Specific Heat-Shock Protein 70 PCRs for Global Leishmania Species Identification. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 1453–1461. [Google Scholar] [CrossRef]
- Del Río, L.; Chitimia, L.; Cubas, A.; Victoriano, I.; De la Rúa, P.; Gerrikagoitia, X.; Barral, M.; Muñoz-García, C.I.; Goyena, E.; García-Martínez, D.; et al. Evidence for Widespread Leishmania infantum Infection among Wild Carnivores in L. infantum Periendemic Northern Spain. Prev. Vet. Med. 2014, 113, 430–435. [Google Scholar] [CrossRef] [PubMed]
- Schönian, G.; Nasereddin, A.; Dinse, N.; Schweynoch, C.; Schallig, H.D.F.H.; Presber, W.; Jaffe, C.L. PCR Diagnosis and Characterization of Leishmania in Local and Imported Clinical Samples. Diagn. Microbiol. Infect. Dis. 2003, 47, 349–358. [Google Scholar] [CrossRef] [PubMed]
- de Almeida, J.V.; de Souza, C.F.; Fuzari, A.A.; Joya, C.A.; Valdivia, H.O.; Bartholomeu, D.C.; Brazil, R.P. Diagnosis and Identification of Leishmania Species in Patients with Cutaneous Leishmaniasis in the State of Roraima, Brazil’s Amazon Region. Parasit. Vectors 2021, 14, 32. [Google Scholar] [CrossRef] [PubMed]
- Ortuño, M.; Latrofa, M.S.; Iborra, M.A.; Pérez-Cutillas, P.; Bernal, L.J.; Risueño, J.; Muñoz, C.; Bernal, A.; Sánchez-Lopez, P.F.; Segovia, M.; et al. Genetic Diversity and Phylogenetic Relationships between Leishmania infantum from Dogs, Humans and Wildlife in South-East Spain. Zoonoses Public Health 2019, 66, 961–973. [Google Scholar] [CrossRef] [PubMed]
- Kuhls, K.; Chicharro, C.; Cañavate, C.; Cortes, S.; Campino, L.; Haralambous, C.; Soteriadou, K.; Pratlong, F.; Dedet, J.P.; Mauricio, I.; et al. Differentiation and Gene Flow among European Populations of Leishmania infantum MON-1. PLoS Negl. Trop. Dis. 2008, 2, e261. [Google Scholar] [CrossRef] [PubMed]
- Tomás-Pérez, M.; Hide, M.; Riera, C.; Montoya, L.; Bañuls, A.L.; Ribera, E.; Portús, M.; Fisa, R. Multilocus Microsatellite Typing of Leishmania infantum Isolates in Monitored Leishmania/HIV Coinfected Patients. Parasit. Vectors 2015, 8, 386. [Google Scholar] [CrossRef] [PubMed]
- Botilde, Y.; Laurent, T.; Quispe Tintaya, W.; Chicharro, C.; Cañavate, C.; Cruz, I.; Kuhls, K.; Schönian, G.; Dujardin, J.C. Comparison of Molecular Markers for Strain Typing of Leishmania infantum. Infect. Genet. Evol. 2006, 6, 440–446. [Google Scholar] [CrossRef] [PubMed]
- Seridi, N.; Belkaid, M.; Quispe-Tintaya, W.; Zidane, C.; Dujardin, J.C. Application of PCR-RFLP for the Exploration of the Molecular Diversity of Leishmania infantum in Algeria. Trans. R. Soc. Trop. Med. Hyg. 2008, 102, 556–563. [Google Scholar] [CrossRef] [PubMed]
- Millán, J.; Zanet, S.; Gomis, M.; Trisciuoglio, A.; Negre, N.; Ferroglio, E. An Investigation into Alternative Reservoirs of Canine Leishmaniasis on the Endemic Island of Mallorca (Spain). Transbound. Emerg. Dis. 2011, 58, 352–357. [Google Scholar] [CrossRef]
- Souza Castro, L.; de Oliveira França, A.; de Castro Ferreira, E.; da Costa Lima Júnior, M.S.; Gontijo, C.M.F.; Pereira, A.A.S.; Dorval, M.E.C. Characterization of Leishmania Species from Central-West Region of Brazil. Parasitol. Res. 2018, 117, 1839–1845. [Google Scholar] [CrossRef]
- Chicharro, C.; Morales, M.A.; Serra, T.; Ares, M.; Salas, A.; Alvar, J. Molecular Epidemiology of Leishmania infantum on the Island of Majorca: A Comparison of Phenotypic and Genotypic Tools. Trans. R. Soc. Trop. Med. Hyg. 2002, 96, S93–S99. [Google Scholar] [CrossRef]
- Sobrino, R.; Ferroglio, E.; Oleaga, A.; Romano, A.; Millan, J.; Revilla, M.; Arnal, M.C.; Trisciuoglio, A.; Gortázar, C. Characterization of Widespread Canine Leishmaniasis among Wild Carnivores from Spain. Vet. Parasitol. 2008, 155, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Salvatore, D.; Di Francesco, A.; Poglayen, G.; Rugna, G.; Santi, A.; Morandi, B.; Baldelli, R. Molecular Characterization of Leishmania infantum Strains by Kinetoplast DNA RFLP-PCR. Vet. Ital. 2016, 52, 71–75. [Google Scholar] [CrossRef]
- Cortes, S.; Rolão, N.; Ramada, J.; Campino, L. PCR as a Rapid and Sensitive Tool in the Diagnosis of Human and Canine Leishmaniasis Using Leishmania donovani s.l.—Specific Kinetoplastid Primers. Trans. R. Soc. Trop. Med. Hyg. 2004, 98, 12–17. [Google Scholar] [CrossRef]
- Kimura, M. A Simple Method for Estimating Evolutionary Rates of Base Substitutions through Comparative Studies of Nucleotide Sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Bandelt, H.J.; Forster, P.; Röhl, A. Median-Joining Networks for Inferring Intraspecific Phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef]
- Clement, M.; Posada, D.; Crandall, K.A. TCS: A Computer Program to Estimate Gene Genealogies. Mol. Ecol. 2000, 9, 1657–1659. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A Software for Comprehensive Analysis of DNA Polymorphism Data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Huson, D.H.; Bryant, D. Application of Phylogenetic Networks in Evolutionary Studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef]
- Reithinger, R.; Dujardin, J.C. Molecular Diagnosis of Leishmaniasis: Current Status and Future Applications. J. Clin. Microbiol. 2007, 45, 21–25. [Google Scholar] [CrossRef]
- El Tai, N.O.; El Fari, M.; Mauricio, I.; Miles, M.A.; Oskam, L.; El Safi, S.H.; Presber, W.H.; Schönian, G. Leishmania donovani: Intraspecific Polymorphisms of Sudanese Isolates Revealed by PCR-Based Analyses and DNA Sequencing. Exp. Parasitol. 2001, 97, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Guerbouj, S.; Victoir, K.; Guizani, I.; Seridi, N.; Nuwayri-Salti, N.; Belkaid, M.; Ben Ismail, R.; Ray, D.L.E.; Dujardin, J.C. Gp63 Gene Polymorphism and Population Structure of Leishmania donovani Complex: Influence of the Host Selection Pressure? Parasitology 2001, 122, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Lewin, S.; Schönian, G.; El Tai, N.; Oskam, L.; Bastien, P.; Presber, W. Strain Typing in Leishmania donovani by Using Sequence-Confirmed Amplified Region Analysis. Int. J. Parasitol. 2002, 32, 1267–1276. [Google Scholar] [CrossRef] [PubMed]
- Quispe-Tintaya, K.W.; Laurent, T.; Decuypere, S.; Hide, M.; Bañ, A.-L.; De Doncker, S.; Rijal, S.; Cañ, C.; Campino, L.; Dujardin, J.C. Fluorogenic Assay for Molecular Typing of the Leishmania donovani Complex: Taxonomic and Clinical Applications. J. Infect. Dis. 2005, 192, 685–692. [Google Scholar] [CrossRef] [PubMed]
- Brewster, S.; Barker, D.C. Analysis of Minicircle Classes in Leishmania (Viannia) Species. Trans. R. Soc. Trop. Med. Hyg. 2002, 91, S55–S63. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, L.; Prina, E.; Lang, T.; Milon, G. Real-Time PCR for Detection and Quantitation of Leishmania in Mouse Tissues. J. Clin. Microbiol. 2002, 40, 1666–1669. [Google Scholar] [CrossRef]
- Da Silva, T.A.M.; Gomes, L.I.; Oliveira, E.; Coura-Vital, W.; Silva, L.D.A.; Pais, F.S.M.; Ker, H.G.; Reis, A.B.; Rabello, A.; Carneiro, M. Genetic Homogeneity among Leishmania(Leishmania) infantum Isolates from Dog and Human Samples in Belo Horizonte Metropolitan Area (BHMA), Minas Gerais, Brazil. Parasit. Vectors 2015, 8, 226. [Google Scholar] [CrossRef] [PubMed]
- Risueño, J.; Ortuño, M.; Pérez-Cutillas, P.; Goyena, E.; Maia, C.; Cortes, S.; Campino, L.; Bernal, L.J.; Muñoz, C.; Arcenillas, I.; et al. Epidemiological and Genetic Studies Suggest a Common Leishmania infantum Transmission Cycle in Wildlife, Dogs and Humans Associated to Vector Abundance in Southeast Spain. Vet. Parasitol. 2018, 259, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Quinnell, R.J.; Courtenay, O. Transmission, Reservoir Hosts and Control of Zoonotic Visceral Leishmaniasis. Parasitology 2009, 136, 1915–1934. [Google Scholar] [CrossRef]
- Devera, R.; Fernandes, O.; Coura, R. Should Trypanosoma cruzi Be Called “Cruzi” Complex? A Review of the Parasite Diversity and the Potential of Selecting Population after in Vitro Culturing and Mice Infection. Mem. Inst. Oswaldo Cruz 2003, 98, 1–12. [Google Scholar] [CrossRef]
- Pacheco, R.S.; Grimaldi, G.; Homen, H.; Morel, C.M. Population Heterogeneity among Clones of New World Leishmania Species. Parasitology 1990, 100, 393–398. [Google Scholar] [CrossRef] [PubMed]
- Bussotti, G.; Piel, L.; Pescher, P.; Domagalska, M.A.; Rajan, K.S.; Cohen-Chalamish, S.; Doniger, T.; Hiregange, D.G.; Myler, P.J.; Unger, R.; et al. Genome Instability Drives Epistatic Adaptation in the Human Pathogen Leishmania. Proc. Nat. Acad. Sci. USA 2021, 118, e2113744118. [Google Scholar] [CrossRef] [PubMed]
- Dumetz, F.; Imamura, H.; Sanders, M.; Seblova, V.; Myskova, J.; Pescher, P.; Vanaerschot, M.; Meehan, C.J.; Cuypers, B.; De Muylder, G.; et al. Modulation of Aneuploidy in Leishmania donovani during Adaptation to Different in Vitro and in Vivo Environments and Its Impact on Gene Expression. mBio 2017, 8, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Bussotti, G.; Gouzelou, E.; Boité, M.C.; Kherachi, I.; Harrat, Z.; Eddaikra, N.; Mottram, J.C.; Antoniou, M.; Christodoulou, V.; Bali, A.; et al. Leishmania Genome Dynamics during Environmental Adaptation Reveal Strain-Specific Differences in Gene Copy Number Variation, Karyotype Instability, and Telomeric Amplification. mBio 2018, 9, 10–1128. [Google Scholar] [CrossRef] [PubMed]

| Variable Nucleotide Positions | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP Genotypes | 26 | 71 | 122 | 127 | 153 | 299 | 308 | 325 | 354 | 367 | 382 | 412 | Ratio (%) |
| KX098509 | A | C | C | — | A | A | A | G | T | A | — | C | |
| G1 | . | . | . | . | . | . | . | . | . | . | . | . | 23 |
| G2 | . | — | . | . | . | . | . | . | . | . | . | . | 8 |
| G12 * | . | . | . | . | G | . | . | . | . | . | . | . | 4 |
| G13 * | . | . | . | . | . | . | G | . | . | . | . | . | 54 |
| G14 * | . | . | . | . | . | G | G | . | . | . | . | . | 4 |
| G15 * | . | . | . | . | . | . | . | . | . | . | G | T | 4 |
| G17 * | G | . | A | T | . | . | . | — | C | G | . | . | 4 |
| S.n. | I.n. | Host | Type of Tissue | SNP Genotype | RFLP Genotype |
|---|---|---|---|---|---|
| 1 | 1 * | Human | Buffy coat | G13 | B |
| 2 | 1 * | Human | Buffy coat | G13 | B |
| 3 | 1 * | Human | Buffy coat | G13 | B |
| 4 | 1 * | Human | Buffy coat | G13 | B |
| 5 | 1 * | Human | Bone marrow | G13 | B |
| 6 | 2 * | Human | Buffy coat | G13 | B |
| 7 | 2 * | Human | Bone marrow | G14 | W |
| 8 | 2 * | Human | Buffy coat | G13 | B |
| 9 | 3 * | Human | Buffy coat | G13 | B |
| 10 | 3 * | Human | Skin | G13 | B |
| 11 | 4 * | Human | Bone marrow | G1 | B |
| 12 | 4 * | Human | Buffy coat | G1 | B |
| 13 | 5 * | Human | Bone marrow | G1 | B |
| 14 | 6 * | Human | Buffy coat | G13 | B |
| 15 | 6 * | Human | Buffy coat | G13 | B |
| 16 | 6 * | Human | Buffy coat | G13 | B |
| 17 | 6 * | Human | Buffy coat | G13 | B |
| 18 | 7 * | Human | Bone marrow | G13 | B |
| 19 | 8 * | Human | Bone marrow | G17 | F |
| 20 | 9 | Dog | Lymph node | G2 | B |
| 21 | 10 | Dog | Lymph node | G2 | B |
| 22 | 11 | Dog | Lymph node | G15 | N |
| 23 | 12 | Dog | Lymph node | G1 | B |
| 24 | 13 | Dog | Lymph node | G1 | B |
| 25 | 14 | Human | Bone marrow | G1 | B |
| 26 | 15 | Human | Bone marrow | G12 | P |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roca-Geronès, X.; Sala, C.; Marteles, D.; Villanueva-Saz, S.; Riera, C.; Alcover, M.M.; Fisa, R. Genetic Variability in Leishmaniasis-Causing Leishmania infantum in Humans and Dogs from North-East Spain. Animals 2024, 14, 1796. https://doi.org/10.3390/ani14121796
Roca-Geronès X, Sala C, Marteles D, Villanueva-Saz S, Riera C, Alcover MM, Fisa R. Genetic Variability in Leishmaniasis-Causing Leishmania infantum in Humans and Dogs from North-East Spain. Animals. 2024; 14(12):1796. https://doi.org/10.3390/ani14121796
Chicago/Turabian StyleRoca-Geronès, Xavier, Clara Sala, Diana Marteles, Sergio Villanueva-Saz, Cristina Riera, Mª Magdalena Alcover, and Roser Fisa. 2024. "Genetic Variability in Leishmaniasis-Causing Leishmania infantum in Humans and Dogs from North-East Spain" Animals 14, no. 12: 1796. https://doi.org/10.3390/ani14121796







