A Study on the 3D Reconstruction Strategy of a Sheep Body Based on a Kinect v2 Depth Camera Array
Abstract
Simple Summary
Abstract
1. Introduction
2. Material and Methods
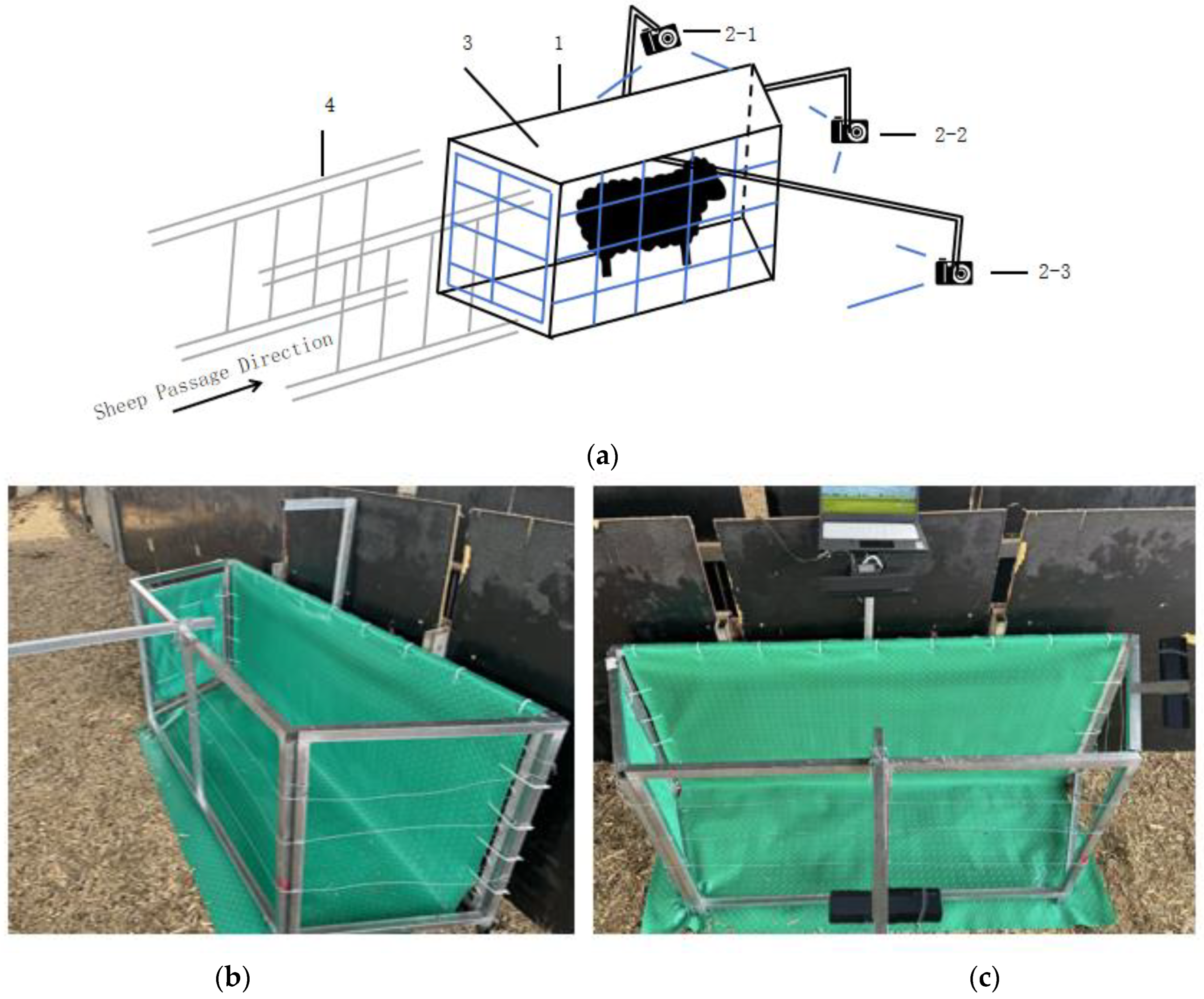
2.1. Design of the Restraining System
2.2. System Parameters
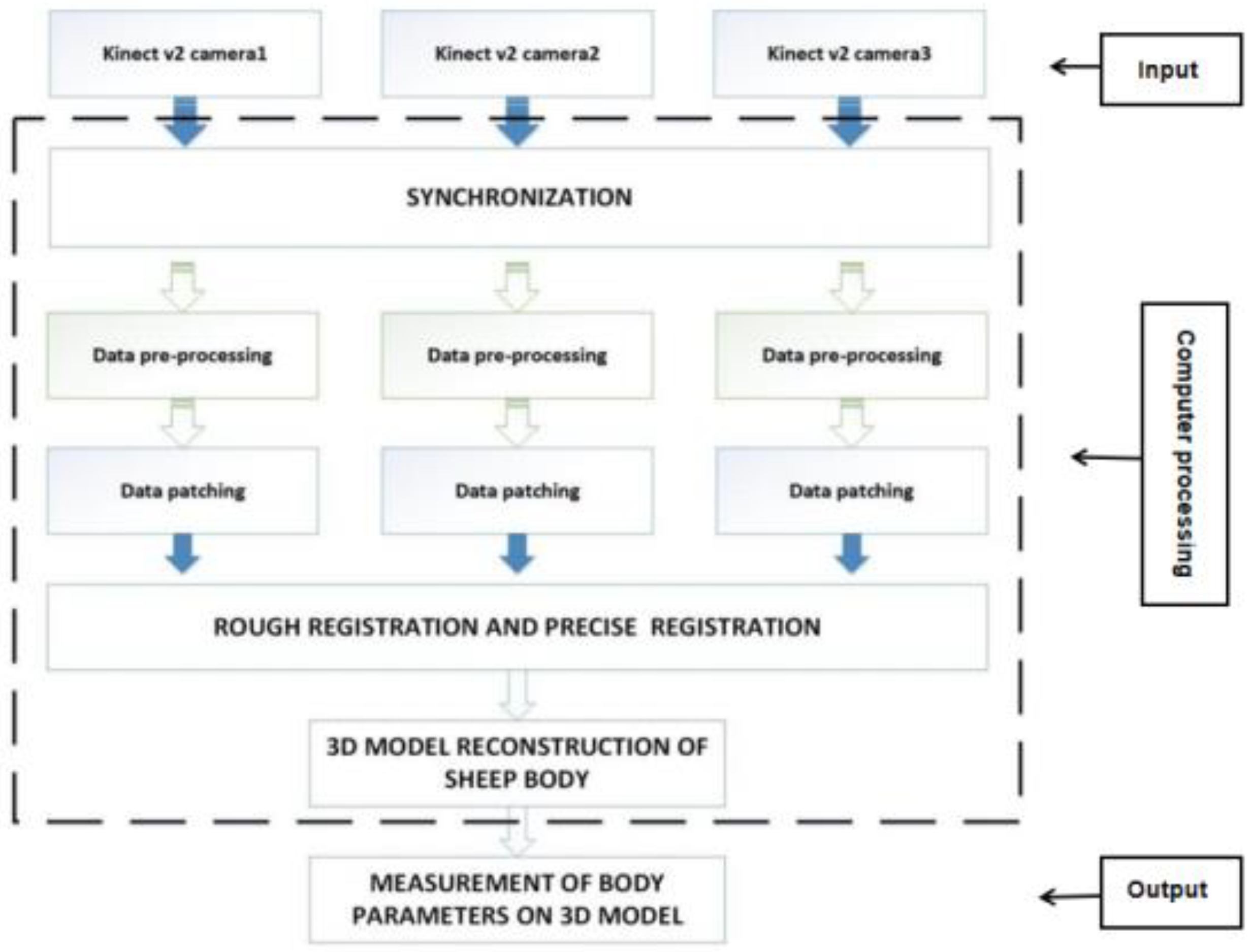
3. Sheep Body Point Cloud Data Acquisition
Data Collection Process
4. 3D Reconstruction of the Sheep Body
4.1. Point Cloud Preprocessing
4.1.1. Point Cloud Denoising
- (1)
- Passthrough Filter: This filter is based on the defined range of points along the x, y, and z axes. It removes unnecessary points to retain the region of interest, thereby obtaining the main body of the sheep within the point cloud channel.
- (2)
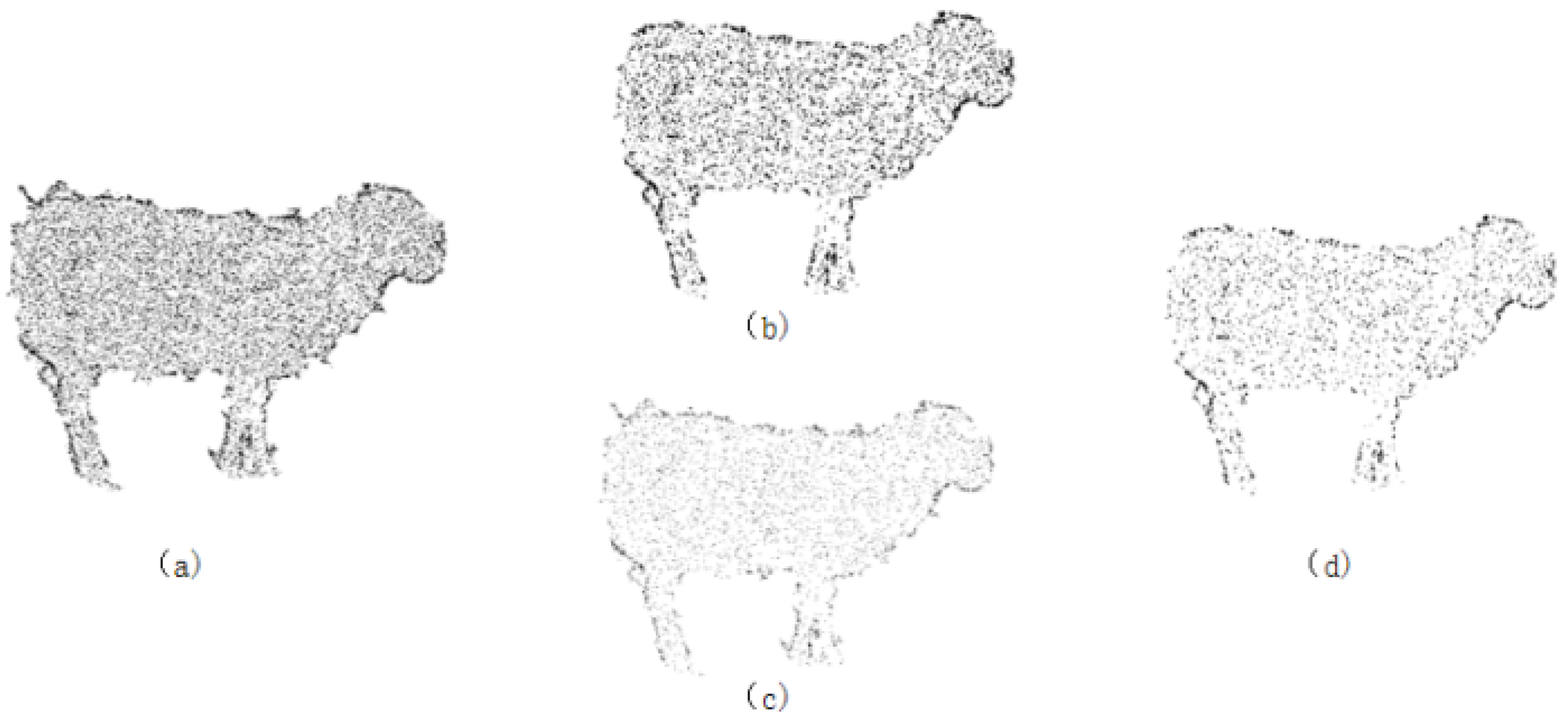
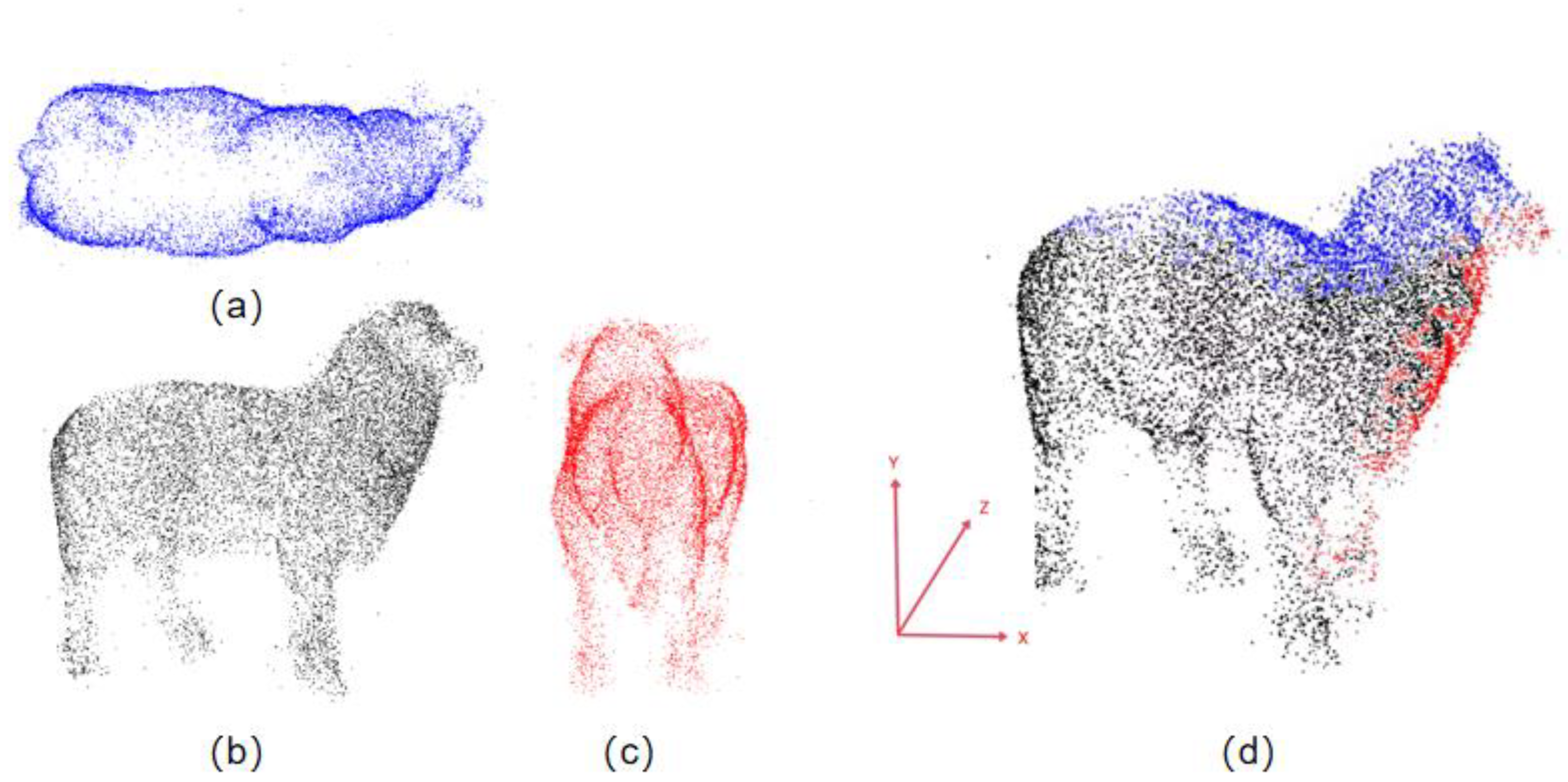
- Statistical Outlier Removal: To eliminate the influence of outlier noise points, statistical filtering is used to remove abnormal points around the sheep, i.e., the elimination of discrete isolated points in the point cloud. This filter is based on the Gaussian distribution characteristics of the points. First, for each point in the point cloud, the k-nearest neighbors are calculated, and the Euclidean distances from these neighbors to the point are computed. The average of these distances is taken as the characteristic distance of the point. Next, a threshold is set based on statistical principles to remove abnormal points: all characteristic distances are statistically analyzed using the mean and standard deviation. A threshold R, which is the mean plus the standard deviation, is set. Points with distances exceeding this threshold are considered abnormal and are removed. In this study, the threshold R is set to 0.6. The effects before and after filtering are shown in Figure 3a,b.
- (3)
- Random Sample Consensus (RANSAC): This algorithm is a ground segmentation algorithm that uses random sampling and iterative optimization of subsets to estimate model parameters. Three data samples are randomly selected from the point cloud data to form a plane. The distance d from any data point in the point cloud to this plane is calculated. If d is less than the distance threshold, the point is considered an inlier, i.e., it lies within the plane. Multiple iterations are performed to remove all outliers from the point cloud data.
- (4)
- After the above filtering processes, the problem of large point cloud data volume still leads to slow computation by the computer. Based on this, this study proposes a conditional voxel filter box, which is the first to combine the advantages of conditional filtering and voxel filtering for animal downsampling to uniformly thin out the point cloud data.
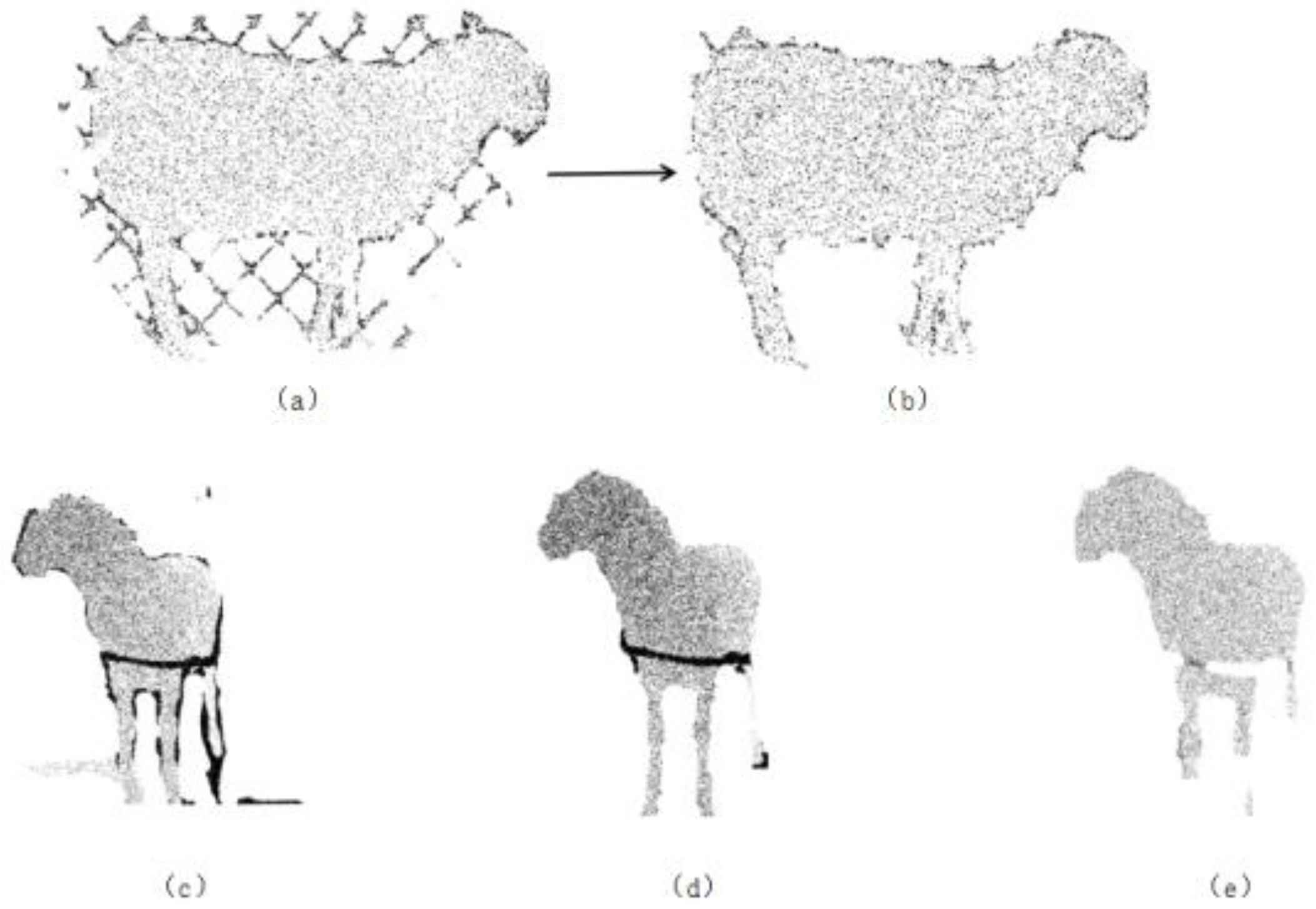
4.1.2. Point Cloud Repair
4.2. 3D Point Cloud Reconstruction from Three Views
4.2.1. RANSAC Algorithm for Coarse Registration
- (1)
- Randomly select non-collinear point pairs: Randomly select three non-collinear corresponding point pairs (F1, F2, F3) from the point set after removing mismatched point pairs.
- (2)
- Solve the transformation matrix: Use the SVD transformation matrix solving method to calculate the transformation matrix HC for F1, F2, and F3.
- (3)
- Calculate the mean squared error: Calculate the mean squared error value K between the source point cloud and the target point cloud.
- (4)
- Compare errors and iterate: Compare the mean squared error value K with the set error threshold V. If K is greater than K, return to step (1) and continue iterating until the error approaches the threshold or the set number of iterations t is reached.
- (5)
- Complete coarse registration: Use the final transformation parameter matrix H to perform coarse registration of the source point cloud with the target point cloud.
4.2.2. ICP Algorithm for Fine Registration
- (1)
- Assume the input point cloud data are P and Q, with an overlapping part . The position of any point in P and Q is PS and QS, respectively. Assume that the initial transformation parameters are RO and TO;
- (2)
- Set the transformation parameters RO and TO and solve for the corresponding point P′ of point cloud P as follows:
- (3)
- Calculate the rigid body transformation matrices R and T to unify the coordinates of the point cloud data.
5. Results and Analysis
5.1. Point Cloud Collection and Reconstruction Results
5.2. Reconstruction Accuracy
6. Discussion
6.1. Impact of Outlier Filtering on Registration
6.2. Discussion on the Effect of a Conditional Voxel Filter Box
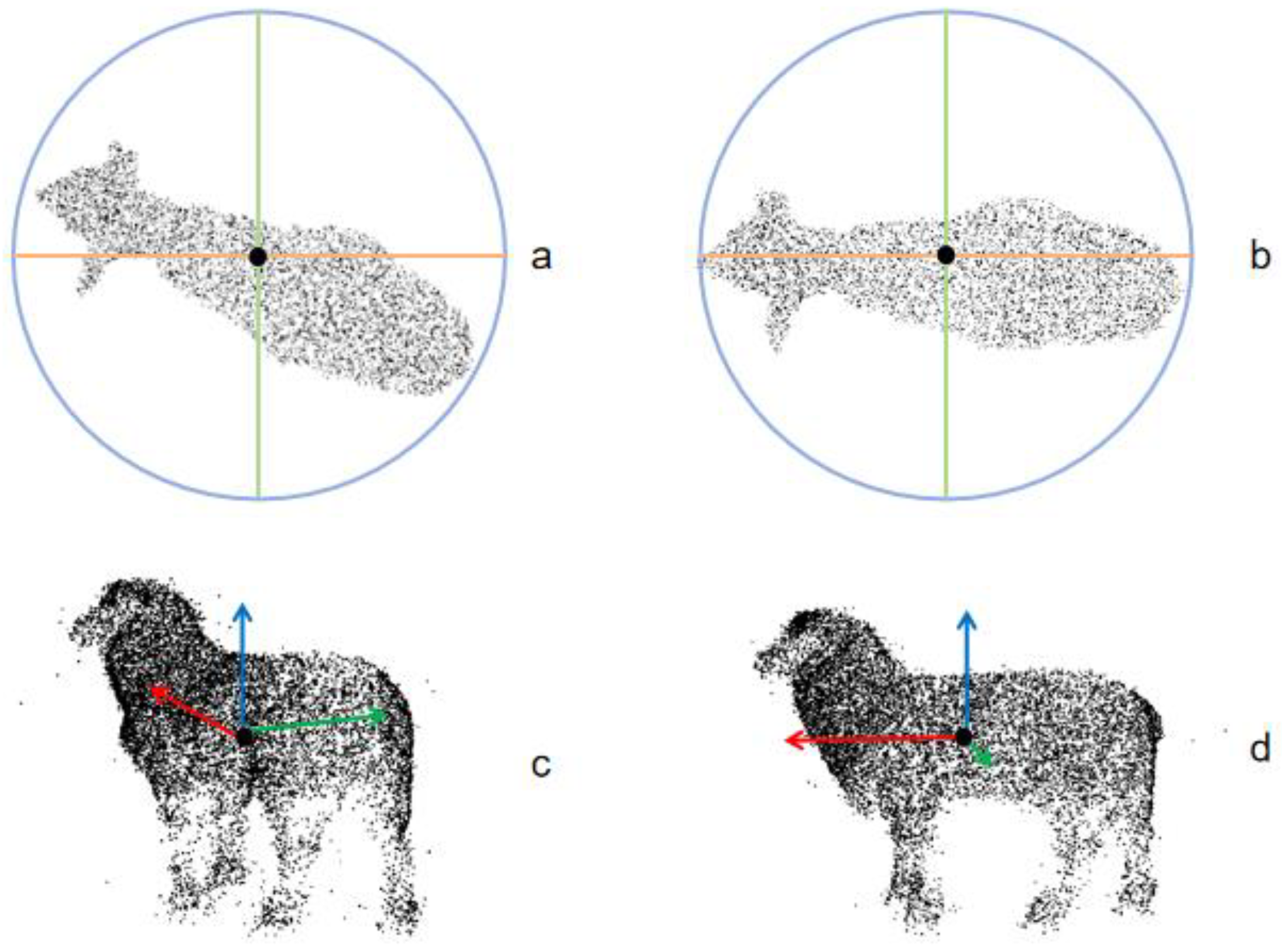
6.3. Discussion on the Effect of Target Rotation Normalization
6.4. Impact of Objective Factors on the Results
- (1)
- The impact of wool length on the measurement error of reconstructed body size. There is a 0.79% error between the software body size measurement of the 3D reconstruction and the manual measurement, mainly due to the impact of wool length on the software measurement. During manual measurement, the wool needs to be compressed to minimize manual error; however, during machine measurement, the point cloud data from the three views are directly registered, resulting in the overall body length data being slightly larger than the manual measurement value. Therefore, further research on body size correction can be conducted in subsequent experiments.
- (2)
- The impact of limiting design on the reconstruction effect. Firstly, the design of the camera angles in the limiting bars is considered. The current design includes frontal, top, and side views, which can basically collect complete point cloud data of the sheep body. However, there may be angle deviations during the registration of some curved surfaces, significantly affecting the 3D reconstruction. When upgrading the limiting bars, adjusting the camera angles or adding more cameras can be considered to collect more comprehensive point cloud data for reconstruction. Secondly, the design of the outer edges of the limiting bars is considered. The current design uses a mesh steel structure, which generates a lot of noise during collection, making preprocessing and registration operations difficult. Therefore, different materials, such as using acrylic panels on one side, can be considered for future improvements to the limiting bars. This would provide unobstructed data collection while limiting and preparing for accurate 3D reconstruction of body size and weight estimation.
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- V’azquez-Arellano, M.; Griepentrog, H.W.; Reiser, D.; Paraforos, D.S. 3-D imaging systems for agricultural applications—A review. Sensors 2018, 16, 618. [Google Scholar] [CrossRef]
- Weinstein, B.G. A computer vision for animal ecology. J. Anim. Ecol. 2018, 87, 533–545. [Google Scholar] [CrossRef]
- Hafid, H. Performances body dimensions of bali cattle of traditional livestock in southeastsulawesi. Indones. J. Agric. Res. 2020, 3, 136–144. [Google Scholar] [CrossRef]
- Michie, C.; Andonovic, I.; Davison, C.; Hamilton, A.; Tachtatzis, C.; Jonsson, N.; Duthie, C.A.; Bowen, J.; Gilroy, M. The Internet of Things enhancing animal welfare and farm operational efficiency. J. Dairy Res. 2020, 87, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Ruchay, A.; Kober, V.; Dorofeev, K.; Kolpakov, V.; Miroshnikov, S. Accurate body measurement of live cattleusing three depth cameras and non-rigid 3-D shape recovery. Comput. Electron. Agric. 2020, 179, 105821. [Google Scholar] [CrossRef]
- Shuai, S.; Ling, Y.; Shihao, L.; Haojie, Z.; Xuhong, T.; Caixing, L.; Aidong, S.; Hanxing, L. Research on 3D surface reconstruction and body size measurement of pigs based on multi-view RGB-D cameras. Comput. Electron. Agric. 2020, 175, 105543. [Google Scholar] [CrossRef]
- Guo, H.; Ma, X.; Ma, Q.; Wang, K.; Su, W.; Zhu, D. LSSA_CAU: An interactive 3d point clouds analysis software for body measurement of livestock with similar forms of cows or pigs. Comput. Electron. Agric. 2017, 138, 60–68. [Google Scholar] [CrossRef]
- Jia, N.; Kootstra, G.; Koerkamp, P.G.; Shi, Z.; Du, S. Segmentation of body parts of cows in RGB-depth images based on template matching. Comput. Electron. Agric. 2021, 180, 105897. [Google Scholar] [CrossRef]
- Tseng, K.H.; Meng-Yun, C.; Jhou, W.H.; Chang, W.J.; Chen-Han, X. Development of Non-Contact Real-Time Monitoring System forAnimal Body Temperature. Proc. Eng. Technol. Innov. 2022, 21, 27–33. [Google Scholar] [CrossRef]
- Neto, C.A.D.; Junior, A.D.S.; Sarmento, J.L.R.; Carvalho, C.G.N.D.; Kridi, D.S.; Carvalho, M.A.M.D. Mobile Device and Software for Automating Body Measurements in Sheep. J. Exp. Agric. Int. 2022, 44, 67–77. [Google Scholar] [CrossRef]
- Becker, S.J.; Fourie, P.J. Phenotypic characterisation of Meatmaster sheep using quantitative and qualitativetrait analysis. Trop. Anim. Health Prod. 2021, 53, 326. [Google Scholar] [CrossRef]
- Tölgyessy, M.; Dekan, M.; Chovanec, Ľ.; Hubinský, P. Evaluation of the azure kinect and its comparison to kinect v1 and kinect v2. Sensors 2021, 21, 413. [Google Scholar] [CrossRef] [PubMed]
- Salem, G.; Krynitsky, J.; Cubert, N.; Pu, A.; Anfinrud, S.; Pedersen, J.; Lehman, J.; Kanuri, A.; Pohida, T. Digital video recorder for Raspberry PI cameras with multi-camera synchronous acquisition. HardwareX 2020, 8, e00160. [Google Scholar] [CrossRef]
- Xiong, X.; Wang, K.; Chen, J.; Li, T.; Lu, B.; Ren, F. A Calibration System of Intelligent Driving Vehicle Mounted Scene Projection Camera Based on Zhang Zhengyou Calibration Method. In Proceedings of the 2022 34th Chinese Controland Decision Conference (CCDC), Hefei, China, 15–17 August 2022; pp. 747–750. [Google Scholar] [CrossRef]
- Du, Q. 3D point cloud registration denoising method for human motion image using deep learning algorithm. Multimed. Syst. 2020, 26, 75–82. [Google Scholar] [CrossRef]
- Hu, W.; Gao, X.; Cheung, G.; Guo, Z. Feature graph learning for 3D point cloud denoising. IEEE Trans. Signal Process. 2020, 68, 2841–2856. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, C.; Ju, Z.; Li, Z.; Su, C.Y. Preprocessing and transmission for 3D point cloud data. In Proceedings of the Intelligent Robotics and Applications: 10th International Conference, ICIRA 2017, Wuhan, China, 16–18 August 2017; Proceedings, Part I 10. Springer International Publishing: Berlin/Heidelberg, Germany, 2017; pp. 438–449. [Google Scholar] [CrossRef]
- Wang, L.; Chen, Y.; Song, W.; Xu, H. Point Cloud Denoising and Feature Preservation: An Adaptive Kernel Approach Based on Local Density and Global Statistics. Sensors 2024, 24, 1718. [Google Scholar] [CrossRef]
- Zhou, L.; Sun, G.; Li, Y.; Li, W.; Su, Z. Point cloud denoising review: From classical to deep learning-based approaches. Graph. Models 2022, 121, 101140. [Google Scholar] [CrossRef]
- Lyu, W.; Ke, W.; Sheng, H.; Ma, X.; Zhang, H. Dynamic Downsampling Algorithm for 3D Point Cloud Map Based on Voxel Filtering. Appl. Sci. 2024, 14, 3160. [Google Scholar] [CrossRef]
- Cao, W.; Wu, J.; Shi, Y.; Chen, D. Restoration of Individual Tree Missing Point Cloud Based on LocalFeatures of Point Cloud. Remote Sens. 2022, 14, 1346. [Google Scholar] [CrossRef]
- Tao, W.; Jiang, G.; Yu, M.; Xu, H.; Song, Y. Point Cloud Projection Based Light-to-Medium G-PCC-1 Hole Distortion Repair Method for Colored Point Cloud. Master’s Thesis, Ningbo University, Ningbo, China, 2023. [Google Scholar] [CrossRef]
- Hong, S.H.; Wang, L.; Truong, T.K. An improved approach to the cubic-spline interpolation. In Proceedings of the 2018 25th IEEE International Conference on Image Processing (ICIP), Athens, Greece, 7–10 October 2018; pp. 1468–1472. [Google Scholar] [CrossRef]
- Abdulmohsin, H.A.; Abdul Wahab, H.B.; Jaber Abdul Hossen, A.M. A Novel Classification Method with Cubic Spline Interpolation. Intell. Autom. Soft Comput. 2022, 31, 339–355. [Google Scholar] [CrossRef]
- Yue, X.; Liu, Z.; Zhu, J.; Gao, X.; Yang, B.; Tian, Y. Coarse-fine point cloud registration based on local point-pair features and the iterative closest point algorithm. Appl. Intell. 2022, 52, 12569–12583. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, H.; Wang, C.; Wang, Y.; Wang, S.; Yang, Z. An Improved RANSAC-ICP Method for Registration of SLAM and UAV-LiDAR Point Cloud at Plot Scale. Forests 2024, 15, 893. [Google Scholar] [CrossRef]
- Keaomanee, Y.; Youngkong, P.; Panpanytep, W.; Maneewarn, T.; Vongbunyong, S. Kinect v2 edgebase registration with ransac-icp. In Proceedings of the 2018 IEEE 4th International Conference on Computer and Communications (ICCC), Chengdu, China, 7–10 December 2018; pp. 1611–1616. [Google Scholar] [CrossRef]
- Li, J.; Ma, W.; Li, Q.; Zhao, C.; Tulpan, D.; Yang, S.; Ding, L.; Gao, R.; Yu, L.; Wang, Z. Multi-view real-time acquisition and 3D reconstruction of point clouds for beef cattle. Comput. Electron. Agric. 2022, 197, 106987. [Google Scholar] [CrossRef]
- Fang, C.; Wu, Z.; Zheng, H.; Yang, J.; Ma, C.; Zhang, T. MCP: Multi-Chicken Pose Estimation Based on Transfer Learning. Animals 2024, 14, 1774. [Google Scholar] [CrossRef]
- Li, J.; Ma, W.; Li, Q.; Xue, X.; Wang, Z. Automatic Acquisition and target extraction of beef cattle 3D point cloud from complex environment. Smart Agric. 2022, 4, 64. [Google Scholar] [CrossRef]
- Zou, B.; Qiu, H.; Lu, Y. Point cloud reduction and denoising based on optimized downsampling and bilateral filtering. IEEE Access 2020, 8, 136316–136326. [Google Scholar] [CrossRef]


| Parameter Category | Value |
|---|---|
| Passage Width × Height (T × H)/(mm) | 600 × 900 |
| Trapezoidal Restraining Pen Width × Height (T × H)/(mm) | 600 × 900 |
| Trapezoidal Restraining Pen Length a (L)/(mm) | 1400 |
| Trapezoidal Restraining Pen Length b (L)/(mm) | 1900 |
| Acute Angle of Trapezoidal Restraining Pen (A)/(°) | 53° |
| Depth Camera Viewing Angle (H × V)/(°) | 70° × 60° |
| Depth Camera Measurement Range (P)/(mm) | 500–4500 |
| Depth Camera Rod Length (L)/(mm) | 900 |
| Serial Number | Tape Measurement (cm) | Point Cloud Measurement (cm) | Error/% | Serial Number | Tape Measurement (cm) | Point Cloud Measurement (cm) | Error/% |
|---|---|---|---|---|---|---|---|
| 1 | 67.1 | 66.0 | 1.7 | 11 | 62.4 | 62.5 | 0.2 |
| 2 | 64.8 | 64.0 | 1.3 | 12 | 64.7 | 64.0 | 1.1 |
| 3 | 67.6 | 67.5 | 0.1 | 13 | 68.1 | 67.5 | 0.9 |
| 4 | 70.4 | 71.0 | 0.8 | 14 | 60.7 | 60.5 | 0.3 |
| 5 | 62.6 | 62.0 | 1.0 | 15 | 66.5 | 66.0 | 0.8 |
| 6 | 70.7 | 70.5 | 0.3 | 16 | 64.9 | 64.0 | 1.4 |
| 7 | 74.1 | 74.0 | 0.1 | 17 | 65.2 | 64.5 | 1.1 |
| 8 | 60.6 | 60.0 | 1.0 | 18 | 63.0 | 62.7 | 0.5 |
| 9 | 62.8 | 62.0 | 1.3 | 19 | 64.3 | 64.0 | 0.5 |
| 10 | 67.3 | 67.0 | 0.4 | 20 | 63.7 | 63.0 | 1.1 |
| Sheep Population | Manual (min) | Average Measurement Speed (min) | Software (min) | Average Measurement Speed (min) | Software (Post-Processing) (min) | Average Measurement Speed (min) |
|---|---|---|---|---|---|---|
| 1 sheep | 7.25 | 7.25 | 1.20 | 1.20 | 0.74 | 0.74 |
| 20 sheep | 138.00 | 6.90 | 24.70 | 1.24 | 14.60 | 0.73 |
| 50 sheep | 317.00 | 6.34 | 67.70 | 1.35 | 36.50 | 0.73 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, J.; Yuan, Z.; Luo, X.; Chen, G.; Wang, C. A Study on the 3D Reconstruction Strategy of a Sheep Body Based on a Kinect v2 Depth Camera Array. Animals 2024, 14, 2457. https://doi.org/10.3390/ani14172457
Liang J, Yuan Z, Luo X, Chen G, Wang C. A Study on the 3D Reconstruction Strategy of a Sheep Body Based on a Kinect v2 Depth Camera Array. Animals. 2024; 14(17):2457. https://doi.org/10.3390/ani14172457
Chicago/Turabian StyleLiang, Jinxin, Zhiyu Yuan, Xinhui Luo, Geng Chen, and Chunxin Wang. 2024. "A Study on the 3D Reconstruction Strategy of a Sheep Body Based on a Kinect v2 Depth Camera Array" Animals 14, no. 17: 2457. https://doi.org/10.3390/ani14172457
APA StyleLiang, J., Yuan, Z., Luo, X., Chen, G., & Wang, C. (2024). A Study on the 3D Reconstruction Strategy of a Sheep Body Based on a Kinect v2 Depth Camera Array. Animals, 14(17), 2457. https://doi.org/10.3390/ani14172457






