Mitochondrial DNA of the Arabian Camel Camelus dromedarius
Abstract
Simple Summary
Abstract
1. Introduction
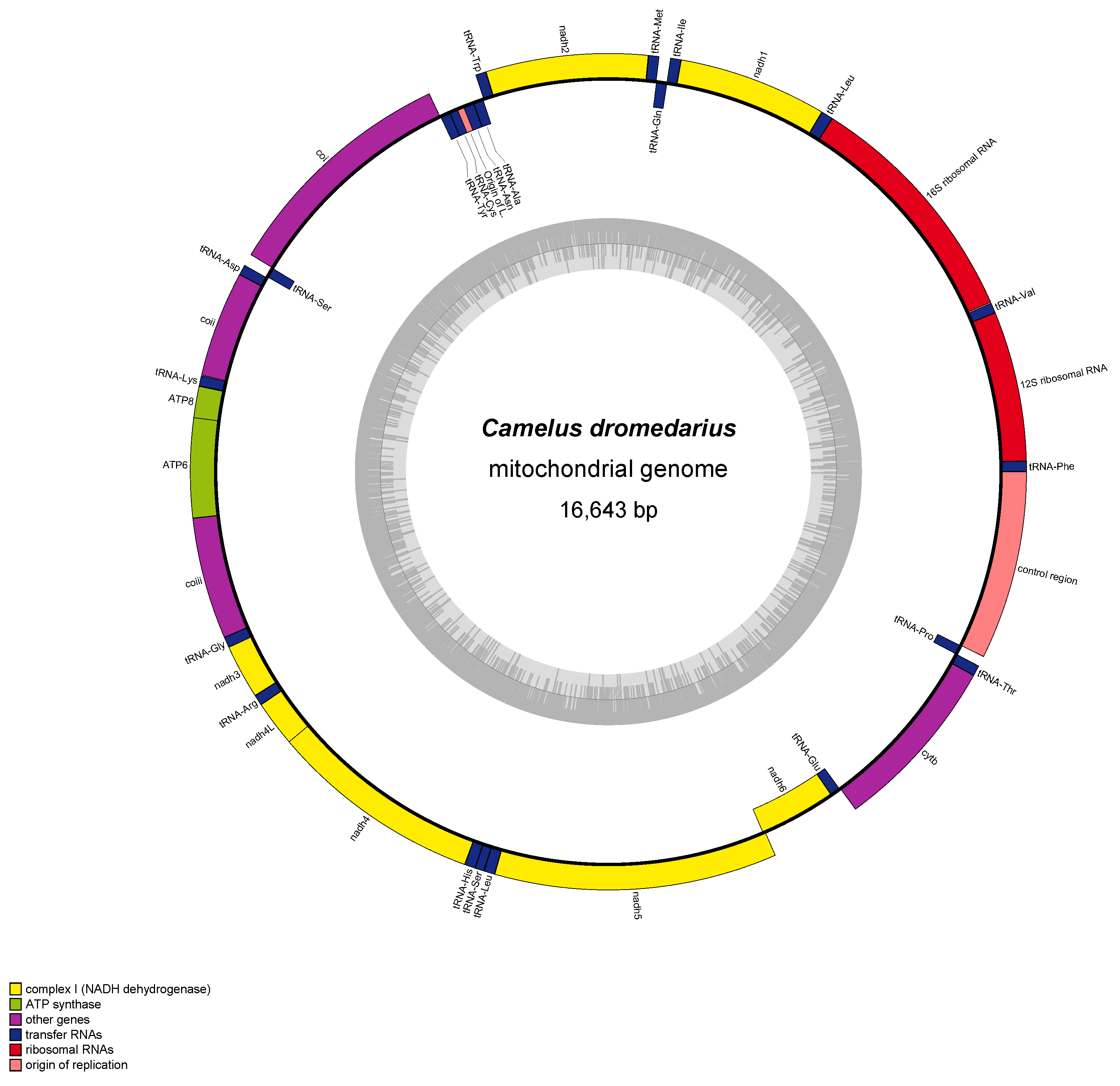
2. Characteristics of Arabian Camel Mitochondrial DNA
3. Mitochondrial Genomes across Various Livestock Species
4. Energy-Related Mitochondrial Genes in the Arabian Camel
5. D-Loop Region of the Arabian Camel Mitogenome
6. Signatures of Natural Selection in the Arabian Camel Mitogenome
7. Genetic Diversity in Arabian Camel Mitochondrial DNA
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mohandesan, E.; Fitak, R.R.; Corander, J.; Yadamsuren, A.; Chuluunbat, B.; Abdelhadi, O.; Raziq, A.; Nagy, P.; Stalder, G.; Walzer, C.; et al. Mitogenome sequencing in the genus Camelus reveals evidence for purifying selection and long-term divergence between wild and domestic bactrian camels. Sci. Rep. 2017, 7, 9970. [Google Scholar] [CrossRef] [PubMed]
- Burger, P.A.; Ciani, E.; Faye, B. Old World camels in a modern world—A balancing act between conservation and genetic improvement. Anim. Genet. 2019, 50, 598–612. [Google Scholar] [CrossRef] [PubMed]
- Heintzman, P.D.; Zazula, G.D.; Cahill, J.A.; Reyes, A.V.; MacPhee, R.D.; Shapiro, B. Genomic data from extinct North American Camelops revise camel evolutionary history. Mol. Biol. Evol. 2015, 32, 2433–2440. [Google Scholar] [CrossRef] [PubMed]
- Pople, A.R.; McLeod, S. Demography of feral camels in central Australia and its relevance to population control. Rangel. J. 2010, 32, 11–19. [Google Scholar] [CrossRef]
- Wu, H.; Guang, X.; Al-Fageeh, M.B.; Cao, J.; Pan, S.; Zhou, H.; Zhang, L.; Abutarboush, M.H.; Xing, Y.; Xie, Z.; et al. Camelid genomes reveal evolution and adaptation to desert environments. Nat. Commun. 2014, 5, 5188. [Google Scholar] [CrossRef]
- Ibrahim, M.A.; Al-Shomrani, B.M.; Simenc, M.; Alharbi, S.N.; Alqahtani, F.H.; Al-Fageeh, M.B.; Manee, M.M. Comparative analysis of transposable elements provides insights into genome evolution in the genus Camelus. BMC Genom. 2021, 22, 842. [Google Scholar] [CrossRef]
- Mahmoud, A.H.; Abu-Tarbush, F.M.; Alshaik, M.; Aljumaah, R.; Saleh, A. Genetic diversity and population genetic structure of six dromedary camel (Camelus dromedarius) populations in Saudi Arabia. Saudi J. Biol. Sci. 2020, 27, 1384–1389. [Google Scholar] [CrossRef]
- Manee, M.M.; Alharbi, S.N.; Algarni, A.T.; Alghamdi, W.M.; Altammami, M.A.; Alkhrayef, M.N.; Alnafjan, B.M. Molecular cloning, bioinformatics analysis, and expression of small heat shock protein beta-1 from Camelus dromedarius, Arabian camel. PLoS ONE 2017, 12, e0189905. [Google Scholar] [CrossRef]
- Satyanarayana, D.S.; Ahlawat, S.; Sharma, R.; Arora, R.; Sharma, A.; Tantia, M.; Vijh, R. Mitochondrial DNA diversity divulges high levels of haplotype diversity and lack of genetic structure in the Indian camels. Gene 2022, 820, 146279. [Google Scholar] [CrossRef]
- Ali, A.; Baby, B.; Vijayan, R. From desert to medicine: A review of camel genomics and therapeutic products. Front. Genet. 2019, 10, 17. [Google Scholar] [CrossRef]
- Ming, L.; Wang, Z.; Yi, L.; Batmunkh, M.; Liu, T.; Siren, D.; He, J.; Juramt, N.; Jambl, T.; Li, Y.; et al. Chromosome-level assembly of wild Bactrian camel genome reveals organization of immune gene loci. Mol. Ecol. Resour. 2020, 20, 770–780. [Google Scholar] [CrossRef] [PubMed]
- Ballard, J.W.O.; Whitlock, M.C. The incomplete natural history of mitochondria. Mol. Ecol. 2004, 13, 729–744. [Google Scholar] [CrossRef] [PubMed]
- Consuegra, S.; John, E.; Verspoor, E.; de Leaniz, C.G. Patterns of natural selection acting on the mitochondrial genome of a locally adapted fish species. Genet. Sel. Evol. 2015, 47, 58. [Google Scholar] [CrossRef] [PubMed]
- van der Valk, T.; Jensen, A.; Caillaud, D.; Guschanski, K. Comparative genomic analyses provide new insights into evolutionary history and conservation genomics of gorillas. BMC Ecol. Evol. 2024, 24, 14. [Google Scholar] [CrossRef]
- Zinovkina, L. Mechanisms of mitochondrial DNA repair in mammals. Biochemistry 2018, 83, 233–249. [Google Scholar] [CrossRef]
- Zhang, L.; Sun, K.; Csorba, G.; Hughes, A.C.; Jin, L.; Xiao, Y.; Feng, J. Complete mitochondrial genomes reveal robust phylogenetic signals and evidence of positive selection in horseshoe bats. BMC Ecol. Evol. 2021, 21, 199. [Google Scholar] [CrossRef]
- Kivisild, T. Maternal ancestry and population history from whole mitochondrial genomes. Investig. Genet. 2015, 6, 3. [Google Scholar] [CrossRef]
- Henn, B.M.; Gignoux, C.R.; Feldman, M.W.; Mountain, J.L. Characterizing the time dependency of human mitochondrial DNA mutation rate estimates. Mol. Biol. Evol. 2009, 26, 217–230. [Google Scholar] [CrossRef]
- Liu, Y.; Dietrich, C.H.; Wei, C. Genetic divergence, population differentiation and phylogeography of the cicada Subpsaltria yangi based on molecular and acoustic data: An example of the early stage of speciation? BMC Evol. Biol. 2019, 19, 5. [Google Scholar] [CrossRef]
- Wollenberg Valero, K.C.; Marshall, J.C.; Bastiaans, E.; Caccone, A.; Camargo, A.; Morando, M.; Niemiller, M.L.; Pabijan, M.; Russello, M.A.; Sinervo, B.; et al. Patterns, mechanisms and genetics of speciation in reptiles and amphibians. Genes 2019, 10, 646. [Google Scholar] [CrossRef]
- Kimura, B.; Marshall, F.B.; Chen, S.; Rosenbom, S.; Moehlman, P.D.; Tuross, N.; Sabin, R.C.; Peters, J.; Barich, B.; Yohannes, H.; et al. Ancient DNA from Nubian and Somali wild ass provides insights into donkey ancestry and domestication. Proc. R. Soc. B Biol. Sci. 2011, 278, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Di Lorenzo, P.; Ceccobelli, S.; Panella, F.; Attard, G.; Lasagna, E. The role of mitochondrial DNA to determine the origin of domestic chicken. World’s Poult. Sci. J. 2015, 71, 311–318. [Google Scholar] [CrossRef]
- Yang, L.; Kong, X.; Yang, S.; Dong, X.; Yang, J.; Gou, X.; Zhang, H. Haplotype diversity in mitochondrial DNA reveals the multiple origins of Tibetan horse. PLoS ONE 2018, 13, e0201564. [Google Scholar] [CrossRef] [PubMed]
- Almathen, F.; Charruau, P.; Mohandesan, E.; Mwacharo, J.M.; Orozco-terWengel, P.; Pitt, D.; Abdussamad, A.M.; Uerpmann, M.; Uerpmann, H.P.; De Cupere, B.; et al. Ancient and modern DNA reveal dynamics of domestication and cross-continental dispersal of the dromedary. Proc. Natl. Acad. Sci. USA 2016, 113, 6707–6712. [Google Scholar] [CrossRef]
- Alaqeely, R.; Alhajeri, B.H.; Almathen, F.; Alhaddad, H. Mitochondrial sequence variation, haplotype diversity, and relationships among dromedary camel-types. Front. Genet. 2021, 12, 723964. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.M.; El-Shazly, S.A.; Sayed, S.M.; Amer, S.A. Molecular study of energy related mitochondrial genes in Arabian and Bactrian camels. Am. J. Biochem. Biotechnol. 2013, 9, 61. [Google Scholar] [CrossRef]
- Stoneking, M.; Hedgecock, D.; Higuchi, R.G.; Vigilant, L.; Erlich, H.A. Population variation of human mtDNA control region sequences detected by enzymatic amplification and sequence-specific oligonucleotide probes. Am. J. Hum. Genet. 1991, 48, 370. [Google Scholar]
- Goldstein, D.; Pollock, D. Launching microsatellites: A review of mutation processes and methods of phylogenetic inference. J. Hered. 1997, 88, 335–342. [Google Scholar] [CrossRef]
- Lei, C.Z.; Chen, H.; Yang, G.S.; Sun, W.B.; Lei, X.Q.; Ge, Q.L.; Wang, Z.F.; Lu, N.; Gao, X.; Hou, W.T. Study on mitochondrial DNA D-loop polymorphism in Chinese donkeys. Yi Chuan Xue Bao Acta Genet. Sin. 2005, 32, 481–486. [Google Scholar]
- Huang, B.; Khan, M.Z.; Chai, W.; Ullah, Q.; Wang, C. Exploring genetic markers: Mitochondrial dna and genomic screening for biodiversity and production traits in donkeys. Animals 2023, 13, 2725. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, J.; Matsumoto, H.; Kim, J.K.; Li, C. Population structure of elongate ilisha Ilisha elongata along the Northwestern Pacific Coast revealed by mitochondrial control region sequences. Fish. Sci. 2016, 82, 771–785. [Google Scholar] [CrossRef]
- Meng, W.; Yang, T.; Hai, S.; Ma, Y.; Cai, L.; Ma, X.; Gao, T.; Guo, Y. Extensive genetic divergence among Diptychus maculatus populations in northwest China. Chin. J. Oceanol. Limnol. 2015, 33, 577–584. [Google Scholar] [CrossRef]
- Van Wijk, R.; Van Solinge, W.W. The energy-less red blood cell is lost: Erythrocyte enzyme abnormalities of glycolysis. Blood 2005, 106, 4034–4042. [Google Scholar] [CrossRef] [PubMed]
- Nunes, M.D.; Dolezal, M.; Schlötterer, C. Extensive paternal mt DNA leakage in natural populations of Drosophila melanogaster. Mol. Ecol. 2013, 22, 2106–2117. [Google Scholar] [CrossRef]
- Kvist, L.; Martens, J.; Nazarenko, A.A.; Orell, M. Paternal leakage of mitochondrial DNA in the great tit (Parus major). Mol. Biol. Evol. 2003, 20, 243–247. [Google Scholar] [CrossRef]
- Clayton, D.A. Replication of animal mitochondrial DNA. Cell 1982, 28, 693–705. [Google Scholar] [CrossRef]
- Amer, S.A. Features of Mitochondrial DNA for The Arabian Camel (Camelus dromedarius). J. Camel Pract. Res. 2015, 22, 235–238. [Google Scholar] [CrossRef]
- Manee, M.M.; Alshehri, M.A.; Binghadir, S.A.; Aldhafer, S.H.; Alswailem, R.M.; Algarni, A.T.; AL-Shomrani, B.M.; AL-Fageeh, M.B. Comparative analysis of camelid mitochondrial genomes. J. Genet. 2019, 98, 88. [Google Scholar] [CrossRef]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Jain, K.; Panigrahi, M.; Nayak, S.S.; Rajawat, D.; Sharma, A.; Sahoo, S.P.; Bhushan, B.; Dutt, T. The evolution of contemporary livestock species: Insights from mitochondrial genome. Gene 2024, 927, 148728. [Google Scholar] [CrossRef]
- Bollongino, R.; Burger, J.; Powell, A.; Mashkour, M.; Vigne, J.D.; Thomas, M.G. Modern taurine cattle descended from small number of near-eastern founders. Mol. Biol. Evol. 2012, 29, 2101–2104. [Google Scholar] [CrossRef]
- Hariyono, D.; Endrawati, E. Indigenous goat genetic resources in Indonesia: Current status and future improvement. J. Adv. Vet. Res. 2023, 13, 141–149. [Google Scholar]
- Hiendleder, S.; Lewalski, H.; Wassmuth, R.; Janke, A. The complete mitochondrial DNA sequence of the domestic sheep (Ovis aries) and comparison with the other major ovine haplotype. J. Mol. Evol. 1998, 47, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Du, L.X.; Ma, Y.H.; Guan, W.J.; Li, H.B.; Zhao, Q.J.; Li, X.; Rao, S.Q. A novel maternal lineage revealed in sheep (Ovis aries). Anim. Genet. 2005, 36, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Tapio, M.; Marzanov, N.; Ozerov, M.; Ćinkulov, M.; Gonzarenko, G.; Kiselyova, T.; Murawski, M.; Viinalass, H.; Kantanen, J. Sheep mitochondrial DNA variation in European, Caucasian, and Central Asian areas. Mol. Biol. Evol. 2006, 23, 1776–1783. [Google Scholar] [CrossRef] [PubMed]
- Larson, G.; Dobney, K.; Albarella, U.; Fang, M.; Matisoo-Smith, E.; Robins, J.; Lowden, S.; Finlayson, H.; Brand, T.; Willerslev, E.; et al. Worldwide phylogeography of wild boar reveals multiple centers of pig domestication. Science 2005, 307, 1618–1621. [Google Scholar] [CrossRef] [PubMed]
- Outram, A.K.; Stear, N.A.; Bendrey, R.; Olsen, S.; Kasparov, A.; Zaibert, V.; Thorpe, N.; Evershed, R.P. The earliest horse harnessing and milking. Science 2009, 323, 1332–1335. [Google Scholar] [CrossRef]
- Cieslak, M.; Pruvost, M.; Benecke, N.; Hofreiter, M.; Morales, A.; Reissmann, M.; Ludwig, A. Origin and history of mitochondrial DNA lineages in domestic horses. PLoS ONE 2010, 5, e15311. [Google Scholar] [CrossRef]
- Cheng, Y.T.; Liu, J.; Yang, L.Q.; Sun, C.; Kong, Q.P. Mitochondrial DNA content contributes to climate adaptation using Chinese populations as a model. PLoS ONE 2013, 8, e79536. [Google Scholar] [CrossRef]
- Mishmar, D.; Ruiz-Pesini, E.; Golik, P.; Macaulay, V.; Clark, A.G.; Hosseini, S.; Brandon, M.; Easley, K.; Chen, E.; Brown, M.D.; et al. Natural selection shaped regional mtDNA variation in humans. Proc. Natl. Acad. Sci. USA 2003, 100, 171–176. [Google Scholar] [CrossRef]
- Bridges, H.R.; Fearnley, I.M.; Hirst, J. The subunit composition of mitochondrial NADH: Ubiquinone oxidoreductase (complex I) from Pichia pastoris. Mol. Cell. Proteom. 2010, 9, 2318–2326. [Google Scholar] [CrossRef] [PubMed]
- DiMauro, S.; Schon, E.A. Mitochondrial respiratory-chain diseases. N. Engl. J. Med. 2003, 348, 2656–2668. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.F.; Kao, C.H.; Chen, Y.T.; Wang, C.H.; Wu, C.Y.; Tsai, C.Y.; Liu, F.C.; Yang, C.W.; Wei, Y.H.; Hsu, M.T.; et al. Cisd2 deficiency drives premature aging and causes mitochondria-mediated defects in mice. Genes Dev. 2009, 23, 1183–1194. [Google Scholar] [CrossRef] [PubMed]
- Romanovsky, Y.M.; Tikhonov, A.N. Molecular energy transducers of the living cell. Proton ATP synthase: A rotating molecular motor. Phys.-Uspekhi 2010, 53, 893. [Google Scholar] [CrossRef]
- Andreu, A.L.; Hanna, M.G.; Reichmann, H.; Bruno, C.; Penn, A.S.; Tanji, K.; Pallotti, F.; Iwata, S.; Bonilla, E.; Lach, B.; et al. Exercise intolerance due to mutations in the cytochrome b gene of mitochondrial DNA. N. Engl. J. Med. 1999, 341, 1037–1044. [Google Scholar] [CrossRef]
- Iyengar, A.; Diniz, F.M.; Gilbert, T.; Woodfine, T.; Knowles, J.; Maclean, N. Structure and evolution of the mitochondrial control region in oryx. Mol. Phylogenet. Evol. 2006, 1, 305–314. [Google Scholar] [CrossRef]
- Shedlock, A.M.; Parker, J.D.; Crispin, D.A.; Pietsch, T.W.; Burmer, G.C. Evolution of the salmonid mitochondrial control region. Mol. Phylogenet. Evol. 1992, 1, 179–192. [Google Scholar] [CrossRef]
- Roques, S.; Godoy, J.A.; Negro, J.J.; Hiraldo, F. Organization and variation of the mitochondrial control region in two vulture species, Gypaetus barbatus and Neophron percnopterus. J. Hered. 2004, 95, 332–337. [Google Scholar] [CrossRef]
- Maté, M.; Di Rocco, F.; Zambelli, A.; Vidal-Rioja, L. Mitochondrial DNA structure and organization of the control region of South American camelids. Mol. Ecol. Notes 2004, 4, 765–767. [Google Scholar] [CrossRef]
- Bahbahani, H.; Al-Zoubi, S.; Ali, F.; Afana, A.; Dashti, M.; Al-Ateeqi, A.; Wragg, D.; Al-Bustan, S.; Almathen, F. Signatures of purifying selection and site-specific positive selection on the mitochondrial DNA of dromedary camels (Camelus dromedarius). Mitochondrion 2023, 69, 36–42. [Google Scholar] [CrossRef]
- Babar, M.E.; Hussain, T.; Wajid, A.; Nawaz, A.; Nadeem, A.; Shah, S.A.; Shahid, M.A.; Ahmad, N.; Javed, K.; Abdullah, M. Mitochondrial Cytochrome-b and D-loop Sequence Based Genetic Diversity in Mareecha and Bareela Camel Breeds of Pakistan. J. Anim. Plant Sci. 2015, 25, 591–594. [Google Scholar]
- Xueqi, W.; Abdussamad, A.M.; Ibrahim, J.; Sanke, O.J.; Olaniyi, W.A.; Dawuda, P.M.; Pan, H.C.; Peng, M.S.; Adeola, A.C.; Zhang, Y.P. Mitochondrial DNA variation of Nigerian dromedary camel (Camelus dromedarius). Anim. Genet. 2021, 52, 570–572. [Google Scholar] [CrossRef]
- Spencer, P.; Woolnough, A. Assessment and genetic characterisation of Australian camels using microsatellite polymorphisms. Livest. Sci. 2010, 129, 241–245. [Google Scholar] [CrossRef]
- Abdussamad, A.; Charruau, P.; Kalla, D.; Burger, P. Validating local knowledge on camels: Colour phenotypes and genetic variation of dromedaries in the Nigeria-Niger corridor. Livest. Sci. 2015, 181, 131–136. [Google Scholar] [CrossRef]
- Legesse, Y.W.; Dunn, C.D.; Mauldin, M.R.; Ordonez-Garza, N.; Rowden, G.R.; Gebre, Y.M.; Kurtu, M.Y.; Mohammed Ali, S.; Whibesilassie, W.D.; Ballou, M.; et al. Morphometric and genetic variation in 8 breeds of Ethiopian camels (Camelus dromedarius). J. Anim. Sci. 2018, 96, 4925–4934. [Google Scholar] [CrossRef] [PubMed]
- Satyanarayana, D.S.; Ahlawat, S.; Sharma, R.; Arora, R.; Sharma, A.; Tantia, M.; Vijh, R. Genetic differentiation of Indian dromedary and Bactrian camel populations based on mitochondrial ATP8 and ATP6 genes. Anim. Biotechnol. 2023, 34, 756–760. [Google Scholar] [CrossRef] [PubMed]
- Hedayat, N.; Khalkhali, R.; Pourasad, K. Investigation of D-loop Region Diversity in Mitochondrial DNA of Iranian Dromedary and Bactrian Camels. Appl. Anim. Sci. Res. J. 2017, 6, 21–24. [Google Scholar]
- Ming, L.; Siren, D.; Yi, L.; Hai, L.; He, J.; Ji, R. Mitochondrial DNA variation and phylogeography of Old World camels. Anim. Biosci. 2021, 34, 525. [Google Scholar] [CrossRef]
- Almathen, F.; Elbir, H.; Bahbahani, H.; Mwacharo, J.; Hanotte, O. Polymorphisms in MC1R and ASIP genes are associated with coat color variation in the Arabian camel. J. Hered. 2018, 109, 700–706. [Google Scholar] [CrossRef]
- Bardakci, F.; Abdelgadir, A.; Alam, M.J.; Biyik, H.H.; Siddiqui, A.J.; Badraoui, R.; Adnan, M.; Alreshidi, M.; Koc, A.; Snoussi, M. A global evaluation of mitochondrial DNA diversity and distribution of dromedary, Camelus dromedarius from north-central Saudi Arabia. J. Genet. 2024, 103, 25. [Google Scholar] [CrossRef]

| Species | Sample Size | Minimum Length | Maximum Length | Mean Length |
|---|---|---|---|---|
| Camelus bactrianus | 135 | 16,398 | 16,856 | 16,617.36 |
| Camelus dromedarius | 125 | 16,375 | 16,738 | 16,620.94 |
| Camelus ferus | 32 | 16,397 | 16,683 | 16,577.78 |
| Camelus knoblochi | 5 | 16,643 | 16,706 | 16,677.80 |
| Lama glama | 1 | 16,084 | 16,084 | 16,084.00 |
| Lama guanicoe | 2 | 16,084 | 16,597 | 16,340.50 |
| Vicugna pacos | 4 | 16,379 | 16,680 | 16,596.25 |
| Vicugna vicugna | 2 | 16,659 | 16,680 | 16,669.50 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manee, M.M.; Al-Shomrani, B.M.; Alqahtani, F.H. Mitochondrial DNA of the Arabian Camel Camelus dromedarius. Animals 2024, 14, 2460. https://doi.org/10.3390/ani14172460
Manee MM, Al-Shomrani BM, Alqahtani FH. Mitochondrial DNA of the Arabian Camel Camelus dromedarius. Animals. 2024; 14(17):2460. https://doi.org/10.3390/ani14172460
Chicago/Turabian StyleManee, Manee M., Badr M. Al-Shomrani, and Fahad H. Alqahtani. 2024. "Mitochondrial DNA of the Arabian Camel Camelus dromedarius" Animals 14, no. 17: 2460. https://doi.org/10.3390/ani14172460
APA StyleManee, M. M., Al-Shomrani, B. M., & Alqahtani, F. H. (2024). Mitochondrial DNA of the Arabian Camel Camelus dromedarius. Animals, 14(17), 2460. https://doi.org/10.3390/ani14172460







