Heat Stress Induces Alterations in Gene Expression of Actin Cytoskeleton and Filament of Cellular Components Causing Gut Disruption in Growing–Finishing Pigs
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Treatments
2.2. Sample Collection
2.3. Transcriptomic Analysis
2.4. Bioinformatic Analysis
2.5. RNA Extraction and Quantitative Real-Time PCR
2.6. Statistical Analysis
3. Results
3.1. Analysis of DEGs
3.2. Clustering and Functional Enrichment Analysis
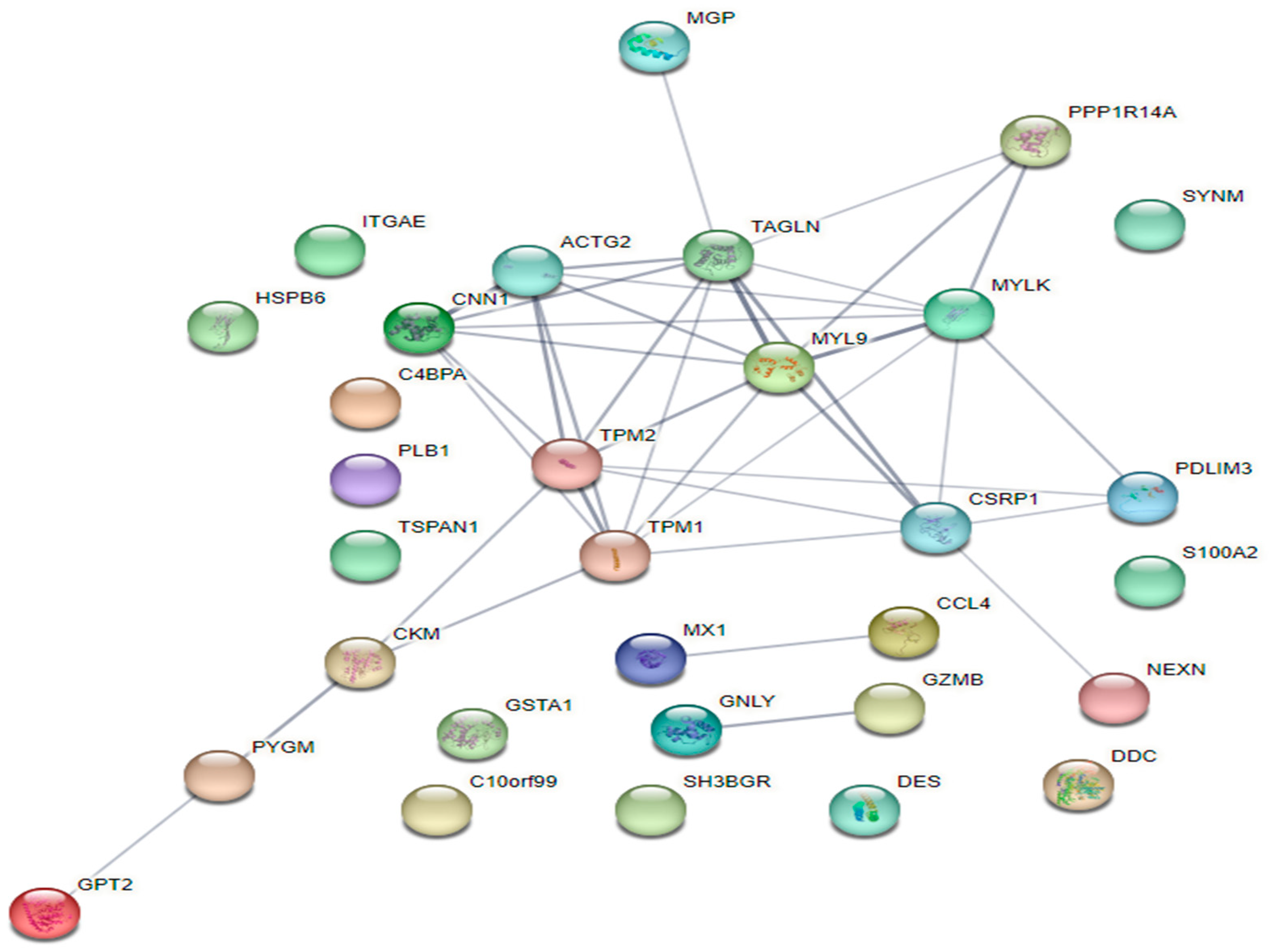
3.3. Protein-Protein Interaction (PPI) Network Analysis
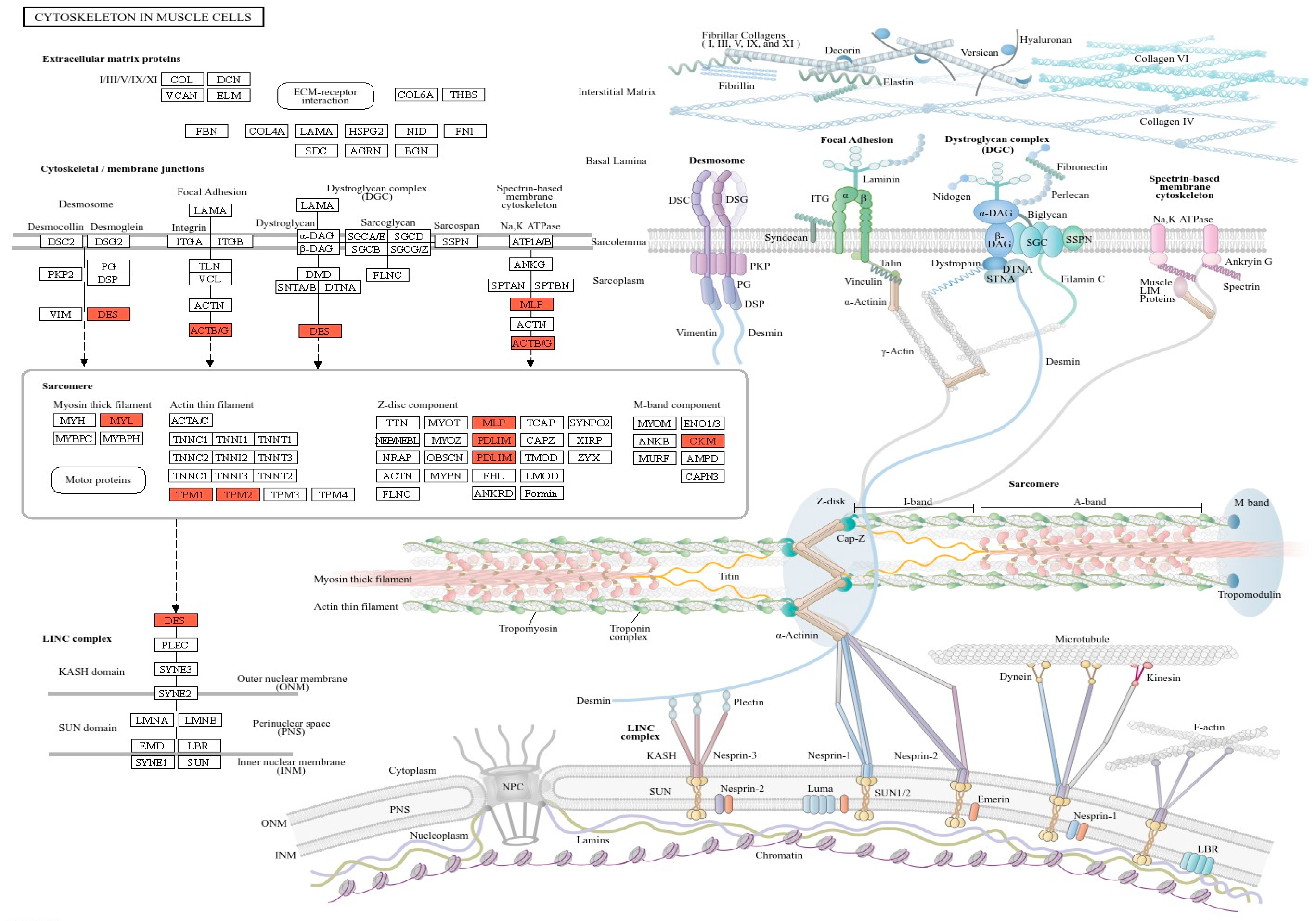
3.4. Key Candidate Genes Involving in Actin Cytoskeleton and Filament Formation Pathway
3.5. Quantitative Real-Time PCR Validation of Gene Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rebez, E.B.; Sejian, V.; Silpa, M.V.; Dunshea, F.R. Heat stress and histopathological changes of vital organs: A novel approach to assess climate resilience in farm animals. Sustainability 2023, 15, 1242. [Google Scholar] [CrossRef]
- Silva, W.C.D.; Silva, J.A.R.D.; Martorano, L.G.; Silva, É.B.R.D.; Sousa, C.E.L.; Neves, K.A.L.; de Araújo, C.V.; Joaquim, L.A.; Rodrigues, T.C.G.D.C.; Belo, T.S.; et al. Thermographic Profiles in Livestock Systems under Full Sun and Shaded Pastures during an Extreme Climate Event in the Eastern Amazon, Brazil: El Niño of 2023. Animals 2024, 14, 855. [Google Scholar] [CrossRef] [PubMed]
- Ghezzi, M.D.; Napolitano, F.; Casas-Alvarado, A.; Hernández-Ávalos, I.; Domínguez-Oliva, A.; Olmos-Hernández, A.; Pereira, A.M. Utilization of infrared thermography in assessing thermal responses of farm animals under heat stress. Animals 2024, 14, 616. [Google Scholar] [CrossRef] [PubMed]
- Mount, L.E. Heat transfer between animal and environment. Proc. Nutr. Soc. 1978, 37, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Cottrell, J.J.; Liu, F.; Hung, A.T.; DiGiacomo, K.; Chauhan, S.S.; Leury, B.J.; Furness, J.B.; Celi, P.; Dunshea, F.R. Nutritional strategies to alleviate heat stress in pigs. Anim. Prod. Sci. 2015, 55, 1391–1402. [Google Scholar] [CrossRef]
- Gabler, N.K.; Pearce, S.C. The impact of heat stress on intestinal function and productivity in grow-finish pigs. Anim. Prod. Sci. 2015, 55, 1403–1410. [Google Scholar] [CrossRef]
- Serviento, A.M.; Labussière, E.; Castex, M.; Renaudeau, D.J.J. Effect of heat stress and feeding management on growth performance and physiological responses of finishing pigs. J. Anim. Sci. 2020, 98, skaa387. [Google Scholar] [CrossRef]
- Lambert, G.P. Stress-induced gastrointestinal barrier dysfunction and its inflammatory effects. J. Anim. Sci. 2009, 87, E101–E108. [Google Scholar] [CrossRef]
- Yan, Y.E.; Zhao, Y.Q.; Wang, H.; Fan, M. Pathophysiological factors underlying heatstroke. Med. Hypotheses 2006, 67, 609–617. [Google Scholar] [CrossRef]
- Patra, A.K.; Kar, I. Heat stress on microbiota composition, barrier integrity, and nutrient transport in gut, production performance, and its amelioration in farm animals. J. Anim. Sci. Technol. 2021, 63, 211–247. [Google Scholar] [CrossRef]
- Pearce, S.C.; Mani, V.; Boddicker, R.L.; Johnson, J.S.; Weber, T.E.; Ross, J.W.; Rhoads, R.P.; Baumgard, L.H.; Gabler, N.K. Heat stress reduces intestinal barrier integrity and favors intestinal glucose transport in growing pigs. PLoS ONE 2013, 8, e70215. [Google Scholar] [CrossRef]
- Turner, J. Intestinal mucosal barrier function in health and disease. Nat. Rev. Immunol. 2009, 9, 799–809. [Google Scholar] [CrossRef]
- Fanning, A.S.; Jameson, B.J.; Jesaitis, L.A.; Anderson, J.M. The tight junction protein ZO-1 establishes a link between the transmembrane protein occludin and the actin cytoskeleton. J. Biol. Chem. 1998, 273, 29745–29753. [Google Scholar] [CrossRef] [PubMed]
- Ulluwishewa, D.; Anderson, R.C.; McNabb, W.C.; Moughan, P.J.; Wells, J.M.; Roy, N.C. Regulation of tight junction permeability by intestinal bacteria and dietary components. J. Nutr. 2011, 141, 769–776. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Roura, E.; Choi, Y.; Kim, J. Transcriptomic analysis of the porcine gut in response to heat stress and dietary soluble fiber from beet pulp. Genes 2022, 13, 1456. [Google Scholar] [CrossRef] [PubMed]
- National Research Council. Nutrient Requirements of Swine, 11th ed.; National Academy Press: Washington, DC, USA, 2012. [Google Scholar]
- A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 15 April 2022).
- FASTX Toolkit. Available online: http://hannonlab.cshl.edu/fastx_toolkit/ (accessed on 15 April 2022).
- BBMap. Available online: https://sourceforge.net/projects/bbmap/ (accessed on 15 April 2022).
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing [Computer Software]. Available online: https://www.R-project.org/ (accessed on 20 November 2020).
- Roberts, A.; Trapnell, C.; Donaghey, J.; Rinn, J.L.; Pachter, L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011, 12, R22. [Google Scholar] [CrossRef]
- Lan, X.; Hsieh, J.C.; Schmidt, C.J.; Zhu, Q.; Lamont, S.J. Liver transcriptome response to hyperthermic stress in three distinct chicken lines. BMC Genom. 2016, 17, 955–966. [Google Scholar] [CrossRef]
- Srikanth, K.; Park, J.E.; Ji, S.Y.; Kim, K.H.; Lee, Y.K.; Kumar, H.; Kim, M.; Baek, Y.C.; Kim, H.; Jang, G.W.; et al. Genome-wide transcriptome and metabolome analyses provide novel insights and suggest a sex-specific response to heat stress in pigs. Genes 2020, 11, 540. [Google Scholar] [CrossRef]
- Sarais, F.; Metzger, K.; Hadlich, F.; Kalbe, C.; Ponsuksili, S. Transcriptomic response of differentiating porcine myotubes to thermal stress and donor piglet age. Int. J. Mol. Sci. 2023, 24, 13599. [Google Scholar] [CrossRef]
- Renaudeau, D.; Collin, A.; Yahav, S.; De Basilio, V.; Gourdine, J.L.; Collier, R.J. Adaptation to hot climate and strategies to alleviate heat stress in livestock production. Animal 2012, 6, 707–728. [Google Scholar] [CrossRef] [PubMed]
- Becker, C.A.; Collier, R.J.; Stone, A.E. Invited review: Physiological and behavioral effects of heat stress in dairy cows. J. Dairy Sci. 2020, 103, 6751–6770. [Google Scholar] [CrossRef]
- Silva, W.C.D.; Silva, J.A.R.D.; Camargo-Júnior, R.N.C.; Silva, É.B.R.D.; Santos, M.R.P.D.; Viana, R.B.; Silva, A.G.M.; Silva, C.M.G.D.; Lourenço-Júnior, J.D.B. Animal welfare and effects of per-female stress on male and cattle reproduction—A review. Front. Vet. Sci. 2023, 10, 1083469. [Google Scholar] [CrossRef] [PubMed]
- Ross, J.W.; Hale, B.J.; Gabler, N.K.; Rhoads, R.P.; Keating, A.F.; Baumgard, L.H. Physiological consequences of heat stress in pigs. Anim. Prod. Sci. 2015, 55, 1381–1390. [Google Scholar] [CrossRef]
- Baumgard, L.H.; Rhoads, R.P. Effects of heat stress on postabsorptive metabolism and energetics. Annu. Rev. Anim. Biosci. 2013, 1, 311–337. [Google Scholar] [CrossRef] [PubMed]
- Godyń, D.; Herbut, P.; Angrecka, S.; Corrêa Vieira, F.M. Use of different cooling methods in pig facilities to alleviate the effects of heat stress—A review. Animals 2020, 10, 1459. [Google Scholar] [CrossRef]
- Ortega, A.D.S.V.; Szabó, C. Adverse effects of heat stress on the intestinal integrity and function of pigs and the mitigation capacity of dietary antioxidants: A review. Animals 2021, 11, 1135. [Google Scholar] [CrossRef]
- Lambert, G.P.; Gisolfi, C.V.; Berg, D.J.; Moseley, P.L.; Oberley, L.W.; Kregel, K.C. Selected contribution: Hyperthermia-induced intestinal permeability and the role of oxidative and nitrosative stress. J. Appl. Physiol. 2002, 92, 1750–1761. [Google Scholar] [CrossRef]
- Ghulam Mohyuddin, S.; Khan, I.; Zada, A.; Qamar, A.; Arbab, A.A.I.; Ma, X.B.; Yu, Z.C.; Liu, X.X.; Yong, Y.H.; Ju, X.H.; et al. Influence of heat stress on intestinal epithelial barrier function, tight junction protein, and immune and reproductive physiology. Biomed. Res. Int. 2022, 2022, 8547379. [Google Scholar] [CrossRef]
- Pearce, S.C.; Mani, V.; Weber, T.E.; Rhoads, R.P.; Patience, J.F.; Baumgard, L.H.; Gabler, N.K. Heat stress and reduced plane of nutrition decreases intestinal integrity and function in pigs. J. Anim. Sci. 2013, 91, 5183–5193. [Google Scholar] [CrossRef]
- Tang, J.; Cao, L.; Jia, G.; Liu, G.; Chen, X.; Tian, G.; Cai, J.; Shang, H.; Zhao, H. The protective effect of selenium from heat stress-induced porcine small intestinal epithelial cell line (IPEC-J2) injury is associated with regulation expression of selenoproteins. Br. J. Nutr. 2019, 122, 1081–1090. [Google Scholar] [CrossRef]
- Fanning, A.S.; Mitic, L.L.; Anderson, J.M. Transmembrane proteins in the tight junction barrier. J. Am. Soc. Nephrol. 1999, 10, 1337–1345. [Google Scholar] [CrossRef] [PubMed]
- Furuse, M.; Fujita, K.; Hiiragi, T.; Fujimoto, K.; Tsukita, S. Claudin-1 and -2: Novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J. Cell Biol. 1998, 141, 1539–1550. [Google Scholar] [CrossRef] [PubMed]
- Furuse, M.; Sasaki, H.; Fujimoto, K.; Tsukita, S. A single gene product, claudin-1 or -2, reconstitutes tight junction strands and recruits occludin in fibroblasts. J. Cell Biol. 1998, 143, 391–401. [Google Scholar] [CrossRef]
- Alizadeh, A.; Akbari, P.; Garssen, J.; Fink-Gremmels, J.; Braber, S. Epithelial integrity, junctional complexes, and biomarkers associated with intestinal functions. Tissue Barriers 2022, 10, 1996830. [Google Scholar] [CrossRef] [PubMed]
- Yong, Y.; Li, J.; Gong, D.; Yu, T.; Wu, L.; Hu, C.; Liu, X.; Yu, Z.; Ma, X.; Gooneratne, R.; et al. ERK1/2 mitogen-activated protein kinase mediates downregulation of intestinal tight junction proteins in heat stress-induced IBD model in pig. J. Therm. Biol. 2021, 101, 103103. [Google Scholar] [CrossRef]
- Yi, H.; Xiong, Y.; Wu, Q.; Wang, M.; Liu, S.; Jiang, Z.; Wang, L. Effects of dietary supplementation with l-arginine on the intestinal barrier function in finishing pigs with heat stress. J. Anim. Physiol. Anim. Nutr. 2020, 104, 1134–1143. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Liu, Y.; Tang, J.; Jia, G.; Liu, G.; Tian, G.; Chen, X.; Cai, J.; Kang, B.; Zhao, H. Selenium exerts protective effects against heat stress-induced barrier disruption and inflammation response in jejunum of growing pigs. J. Sci. Food Agric. 2022, 102, 496–504. [Google Scholar] [CrossRef]
- Ivanov, A.I.; Parkos, C.A.; Nusrat, A. Cytoskeletal regulation of epithelial barrier function during inflammation. Am. J. Pathol. 2010, 177, 512–524. [Google Scholar] [CrossRef]
- Gordon, J.I.; Hermiston, M.L. Differentiation and self-renewal in the mouse gastrointestinal epithelium. Curr. Opin. Cell Biol. 1994, 6, 795–803. [Google Scholar] [CrossRef]
- Thorson, W.; Diaz-Horta, O.; Foster, J.N.; Spiliopoulos, M.; Quintero, R.; Farooq, A.; Blanton, S.; Tekin, M. De novo ACTG2 mutations cause congenital distended bladder, microcolon, and intestinal hypoperistalsis. Hum. Genet. 2014, 133, 737–742. [Google Scholar] [CrossRef] [PubMed]
- Rzeszotek, S.; Trybek, G.; Tarnowski, M.; Serwin, K.; Jaroń, A.; Schneider, G.; Kolasa, A.; Wiszniewska, B. Colostrum-induced temporary changes in the expression of proteins regulating the epithelial barrier function in the intestine. Foods 2022, 11, 685. [Google Scholar] [CrossRef]
- Ok, M.; Yildiz, R.; Hatipoglu, F.; Baspinar, N.; Ider, M.; Üney, K.; Ertürk, A.; Durgut, M.K.; Terzi, F. Use of intestine-related biomarkers for detecting intestinal epithelial damage in neonatal calves with diarrhea. Am. J. Vet. Res. 2020, 81, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Gu, X. Proteomic changes of the porcine small intestine in response to chronic heat stress. J. Mol. Endocrinol. 2015, 55, 277–293. [Google Scholar] [CrossRef]
- Nighot, M.; Al-Sadi, R.; Guo, S.; Rawat, M.; Nighot, P.; Watterson, M.D.; Ma, T.Y. Lipopolysaccharide-induced increase in intestinal epithelial tight permeability is mediated by toll-like receptor 4/myeloid differentiation primary response 88 (MyD88) activation of myosin light chain kinase expression. Am. J. Pathol. 2017, 187, 2698–2710. [Google Scholar] [CrossRef] [PubMed]
- Nighot, M.; Rawat, M.; Al-Sadi, R.; Castillo, E.F.; Nighot, P.; Ma, T.Y. Lipopolysaccharide-induced increase in intestinal permeability is mediated by TAK-1 activation of IKK and MLCK/MYLK gene. Am. J. Pathol. 2019, 189, 797–812. [Google Scholar] [CrossRef]
- Niu, X.; Hu, C.; Chen, S.; Wen, J.; Liu, X.; Yong, Y.; Yu, Z.; Ma, X.; Li, C.; Wardo, M.; et al. Chitosan-gentamicin conjugate attenuates heat stress-induced intestinal barrier injury via the TLR4/STAT6/MYLK signaling pathway: In vitro and in vivo studies. Carbohydr. Polym. 2023, 321, 121279. [Google Scholar] [CrossRef] [PubMed]
- Broschat, K.O.; Burgess, D.R. Low Mr tropomyosin isoforms from chicken brain and intestinal epithelium have distinct actin-binding properties. J. Biol. Chem. 1986, 261, 13350–13359. [Google Scholar] [CrossRef]
- Xu, Y.; Qian, H.; Feng, X.; Xiong, Y.; Lei, M.; Ren, Z.; Zuo, B.; Xu, D.; Ma, Y.; Yuan, H. Differential proteome and transcriptome analysis of porcine skeletal muscle during development. J. Proteom. 2012, 75, 2093–2108. [Google Scholar] [CrossRef]
- Shi, X.; Li, C.; Cao, M.; Xu, X.; Zhou, G.; Xiong, Y.L. Comparative proteomic analysis of longissimus dorsi muscle in immuno- and surgically castrated male pigs. Food Chem. 2016, 199, 885–892. [Google Scholar] [CrossRef]
- Bao, E.; Sultan, K.R.; Nowak, B.; Hartung, J. Localization of heat shock proteins and histopathological changes in the kidneys of transported pigs. Livest. Sci. 2008, 118, 231–237. [Google Scholar] [CrossRef]
- Hao, Y.; Liu, J.R.; Zhang, Y.; Yang, P.G.; Feng, Y.J.; Cui, Y.J.; Yang, C.H.; Gu, X.H. The microRNA expression profile in porcine skeletal muscle is changed by constant heat stress. Anim. Genet. 2016, 47, 365–369. [Google Scholar] [CrossRef] [PubMed]
- Sui, B.; Chen, D.; Liu, W.; Tian, B.; Lv, L.; Pei, J.; Wu, Q.; Zhou, M.; Fu, Z.F.; Zhang, Y.; et al. Comparison of lncRNA and mRNA expression in mouse brains infected by a wild-type and a lab-attenuated Rabies lyssavirus. J. Gen. Virol. 2021, 102, 001538. [Google Scholar] [CrossRef]
- Seo, B.J.; Choi, J.; La, H.; Habib, O.; Choi, Y.; Hong, K.; Do, J.T. Role of mitochondrial fission-related genes in mitochondrial morphology and energy metabolism in mouse embryonic stem cells. Redox Biol. 2020, 36, 101599. [Google Scholar] [CrossRef] [PubMed]
- Stark, R.; Kibbey, R.G. The mitochondrial isoform of phosphoenolpyruvate carboxykinase (PEPCK-M) and glucose homeostasis: Has it been overlooked? Biochim. Biophys. Acta 2014, 1840, 1313–1330. [Google Scholar] [CrossRef]
- Hanson, R.W.; Patel, Y.M. Phosphoenolpyruvate carboxykinase (GTP): The gene and the enzyme. Adv. Enzymol. Relat. Areas Mol. Biol. 1994, 69, 203–281. [Google Scholar]
- Qi, M.; Tan, B.; Wang, J.; Li, J.; Liao, S.; Yan, J.; Liu, Y.; Yin, Y. Small intestinal transcriptome analysis revealed changes of genes involved in nutrition metabolism and immune responses in growth retardation piglets. J. Anim. Sci. 2019, 97, 3795–3808. [Google Scholar] [CrossRef]
- Basu Ball, W.; Neff, J.K.; Gohil, V.M. The role of nonbilayer phospholipids in mitochondrial structure and function. FEBS Lett. 2018, 592, 1273–1290. [Google Scholar] [CrossRef]
- Leithner, K.; Triebl, A.; Trötzmüller, M.; Hinteregger, B.; Leko, P.; Wieser, B.I.; Grasmann, G.; Bertsch, A.L.; Züllig, T.; Stacher, E.; et al. The glycerol backbone of phospholipids derives from noncarbohydrate precursors in starved lung cancer cells. Proc. Natl. Acad. Sci. USA 2018, 115, 6225–6230. [Google Scholar] [CrossRef]
- Cai, P.; Zheng, H.; She, J.; Feng, N.; Zou, H.; Gu, J.; Yuan, Y.; Liu, X.; Liu, Z.; Bian, J. Molecular mechanism of aflatoxin-induced hepatocellular carcinoma derived from a bioinformatics analysis. Toxins 2020, 12, 203. [Google Scholar] [CrossRef]
- Xue, D.; Cheng, Y.; Pang, T.; Kuai, Y.; An, Y.; Wu, K.; Li, Y.; Lai, M.; Wang, B.; Wang, S. Sodium butyrate alleviates deoxynivalenol-induced porcine intestinal barrier disruption by promoting mitochondrial homeostasis via PCK2 signaling. J. Hazard. Mater. 2023, 459, 132013. [Google Scholar] [CrossRef] [PubMed]


| Gene | Primer Sequence (5′-3′) | Product Size (bp) | GenBank No. |
|---|---|---|---|
| ZO1 | F GGGGCAATCTCAACTCCTGT R GGTTGTCCAACTTGGGCAT | 137 | XM 021098896.1 |
| CLDN1 | F TTTCCTCAATACAGGAGGGAAGC R CCCTCTCCCCACATTCGAG | 74 | NM_001244539.1 |
| CLDN3 | F GCCAAGATCCTCTACTCCGC R GAGAGCTGCCTAGCATCTGG | 197 | NM_001160075.1 |
| CLDN4 | F CTCTCGGACACCTTCCCAAG R GCAGTGGGGAAGGTCAAAGG | 192 | XM_005661969.2 |
| OCLN | F TCTCAGCCAGCGTATTCTTTC R GCACATCACGATAACGAGCAT | 111 | XM 005672525.3 |
| ACTG2 | F CCTGGCATTGCCGACAGGAT R GGCCAGGATAGAGCCTCCGA | 123 | XM_021087371.1 |
| DES | F TTCCGAGCGGATGTGGATGC R CTGAAGCTGGGCCTGCAGTT | 135 | NM_001001535.1 |
| PYGM | F GTGGAGATGGCGGAAGAGGC R TCGATGACGTGCCGAAGCTC | 140 | XM_003122588.5 |
| MYL9 | F CACCAAGAAGCGGCCACAGA R TGCCCTCCAGGTACTCGTCC | 188 | NM_001244472.1 |
| MYLK | F ATCAGGGAGTCCCGCCACTT R CTGCTGTGCAGGTGGCTTCT | 142 | XM_001929078.6 |
| PPP1R14A | F TGAGCAAGCTGCAGTCTCCG R CGGTACAGCTCCTCCAAGCG | 163 | NM_214337.1 |
| TPM1 | F TGAGCTGGTGTCGCTGCAAA R ATCGGCTTCAGCATCGGTGG | 130 | NM_001097483.2 |
| TPM2 | F GATGTGGCCTCCCTGAACCG R TTCTCGGCCTCCTCCAGCTT | 103 | NM_001129947.1 |
| CNN1 | F GCAGGAGCAGGAGCTTCGAG R CTGGTGCCAGTTCTGGGTGG | 166 | NM_213878.1 |
| PDLIM3 | F GCTGGCGGCACTCAGAAGAT R TTCGCAGTACAGCTCCCCCT | 174 | NM_001001637.1 |
| PDLIM7 | F AGCAGAATGGACAGCCGCTC R GAGGATGCGGAAGGAACGGG | 122 | XM_021084391.1 |
| PCP4 | F GCTGGGGCCACCAATGGAAA R TGAGACTGAATGGCCACCGC | 128 | XM_021071001.1 |
| PCK1 | F TGCTCCCGGAACCTCTGTGA R GGTCGATGATGGGGCACTGG | 107 | NM_001123158.1 |
| PCK2 | F TCTTTCGGCAGCGGCTATGG R ACATAGCGCTTCTTCCCCGC | 152 | NM_001161753.1 |
| GAPDH | F CCACGGTCCATGCCATCACT R GCCTGCTTCACCACCTTCTTG | 268 | XM_021091114.1 |
| Gene ID | Gene Name | Universal Gene Name |
|---|---|---|
| 100736682 | ZO1 | zonula occludens-1 |
| 100625166 | CLDN1 | claudin 1 |
| 431781 | CLDN3 | claudin 3 |
| 733578 | CLDN4 | claudin 4 |
| 397236 | OCLN | occludin |
| 100520667 | ACTG2 | actin gamma 2, smooth muscle |
| 396725 | DES | desmin |
| 733659 | PYGM | glycogen phosphorylase |
| 100157760 | MYL9 | myosin light chain 9 |
| 396848 | MYLK | myosin light chain kinase |
| 397610 | PPP1R14A | protein phosphatase 1 regulatory inhibitor subunit 14A |
| 100037999 | TPM1 | tropomyosin 1 |
| 396693 | TPM2 | tropomyosin 2 |
| 396911 | CNN1 | calponin 1 |
| 414421 | PDLIM3 | PDZ and LIM domain 3 |
| 100624749 | PDLIM7 | PDZ and LIM domain 7 |
| 110256491 | PCP4 | Purkinje cell protein 4 |
| 100144531 | PCK1 | phosphoenolpyruvate carboxykinase 1 |
| 403165 | PCK2 | phosphoenolpyruvate carboxykinase 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, Y.; Park, H.; Kim, J.; Lee, H.; Kim, M. Heat Stress Induces Alterations in Gene Expression of Actin Cytoskeleton and Filament of Cellular Components Causing Gut Disruption in Growing–Finishing Pigs. Animals 2024, 14, 2476. https://doi.org/10.3390/ani14172476
Choi Y, Park H, Kim J, Lee H, Kim M. Heat Stress Induces Alterations in Gene Expression of Actin Cytoskeleton and Filament of Cellular Components Causing Gut Disruption in Growing–Finishing Pigs. Animals. 2024; 14(17):2476. https://doi.org/10.3390/ani14172476
Chicago/Turabian StyleChoi, Yohan, Hyunju Park, Joeun Kim, Hyunseo Lee, and Minju Kim. 2024. "Heat Stress Induces Alterations in Gene Expression of Actin Cytoskeleton and Filament of Cellular Components Causing Gut Disruption in Growing–Finishing Pigs" Animals 14, no. 17: 2476. https://doi.org/10.3390/ani14172476
APA StyleChoi, Y., Park, H., Kim, J., Lee, H., & Kim, M. (2024). Heat Stress Induces Alterations in Gene Expression of Actin Cytoskeleton and Filament of Cellular Components Causing Gut Disruption in Growing–Finishing Pigs. Animals, 14(17), 2476. https://doi.org/10.3390/ani14172476








