RNA-Seq Analysis of Glycolysis Regulation of Avian Leukosis Virus Subgroup J Replication
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. ALV-J-Infected Treatment
2.3. 2-Deoxy-d-Glucose Treatment
2.4. Glucose Uptake Assay
2.5. Pyruvate Content Assay
2.6. Lactate Content Assay
2.7. Total RNA Extraction, cDNA Synthesis, and Real-Time Quantitative PCR (qRT-PCR)
2.8. CCK-8 Assay
2.9. Western Blot Assay
2.10. RNA Extraction, Library Construction, Sequencing, and Data Quality Control
2.11. Identification and Differential Expression Analysis of circRNAs
2.12. Target Gene Prediction and Functional Enrichment Analysis of Differential circRNAs
2.13. Validation of DE circRNAs
2.14. Statistical Analysis
3. Results
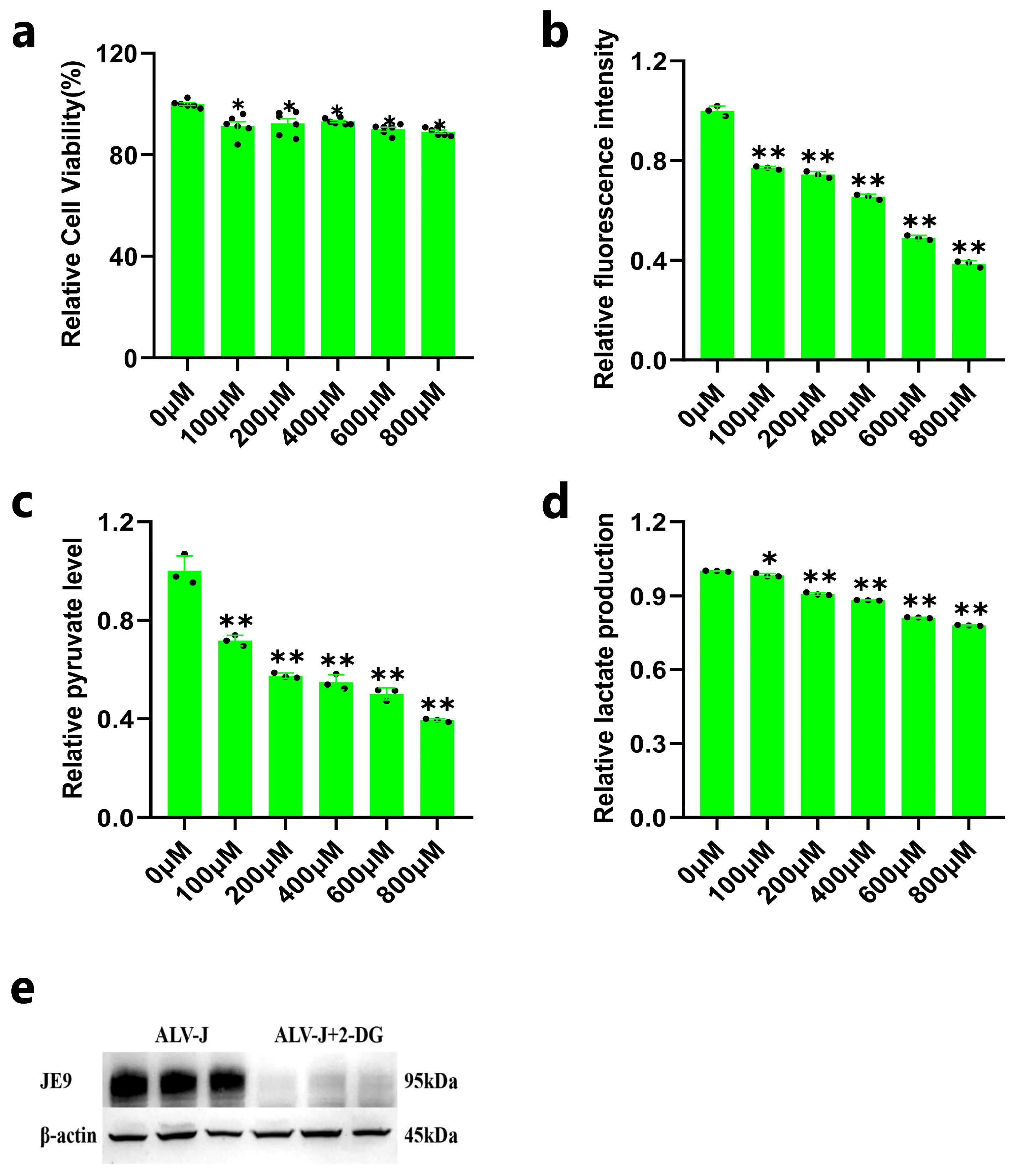
3.1. ALV-J Elevates Glucose Uptake and Glycolysis in DF1 Cells
3.2. Inhibition of DF1 Cell Glycolysis Reduces ALV-J Replication
3.3. Overview of RNA Sequencing Data and Identification of Circular RNAs
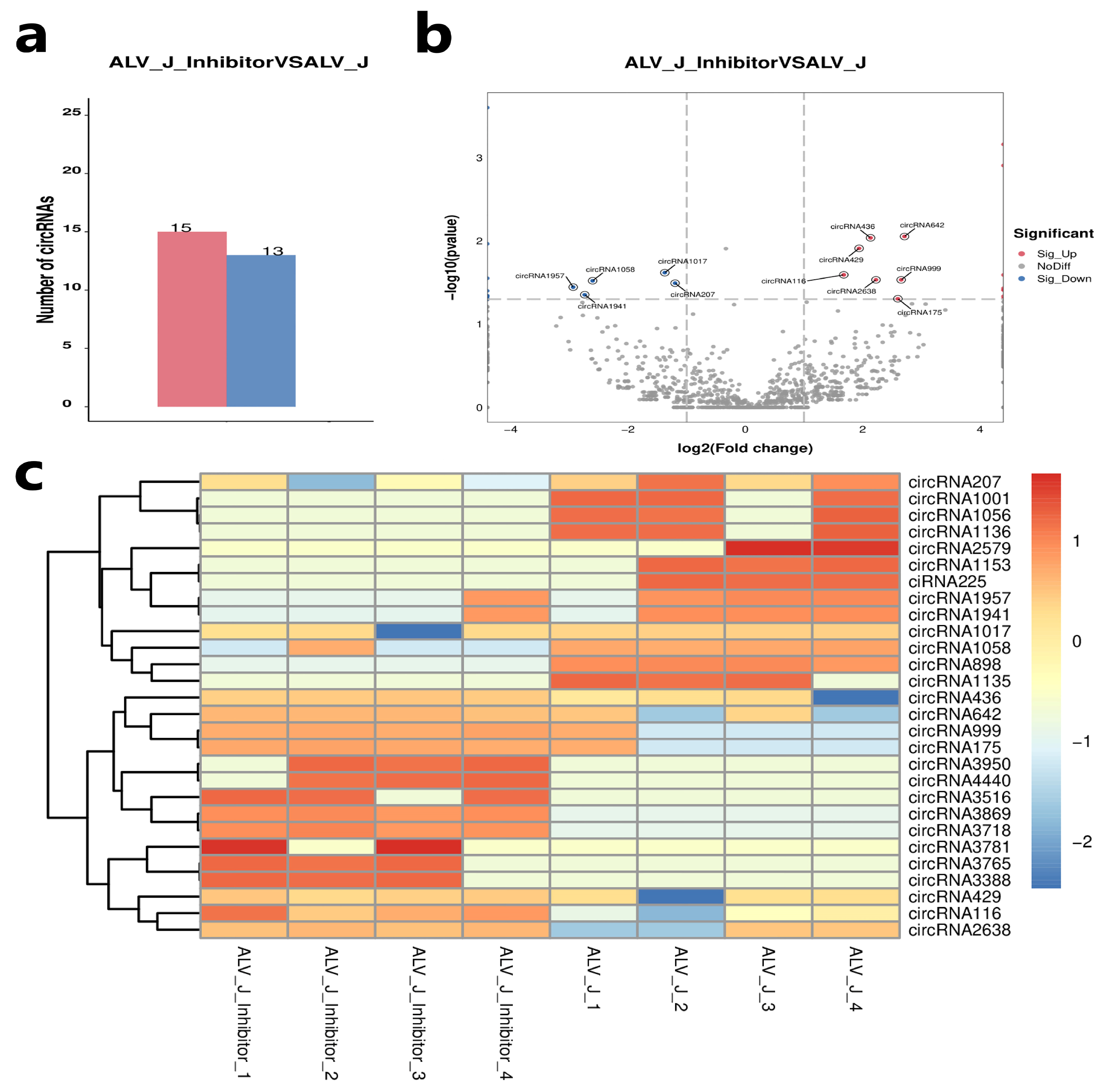
3.4. Identification of Differentially Expressed Circular RNAs
3.5. Validation of Differentially Expressed Circular RNAs
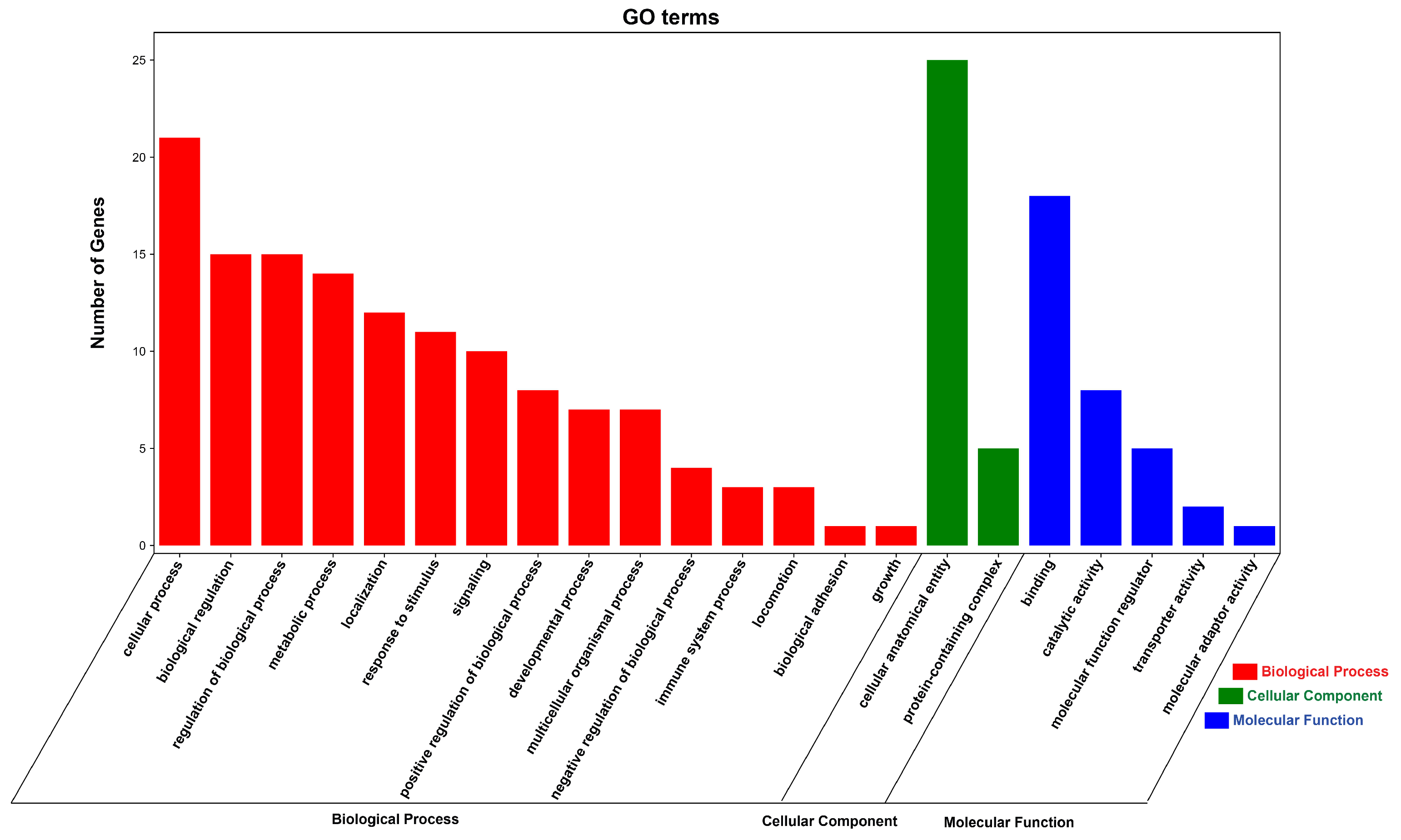
3.6. Functional Enrichment Analysis of DE circRNA Source Genes
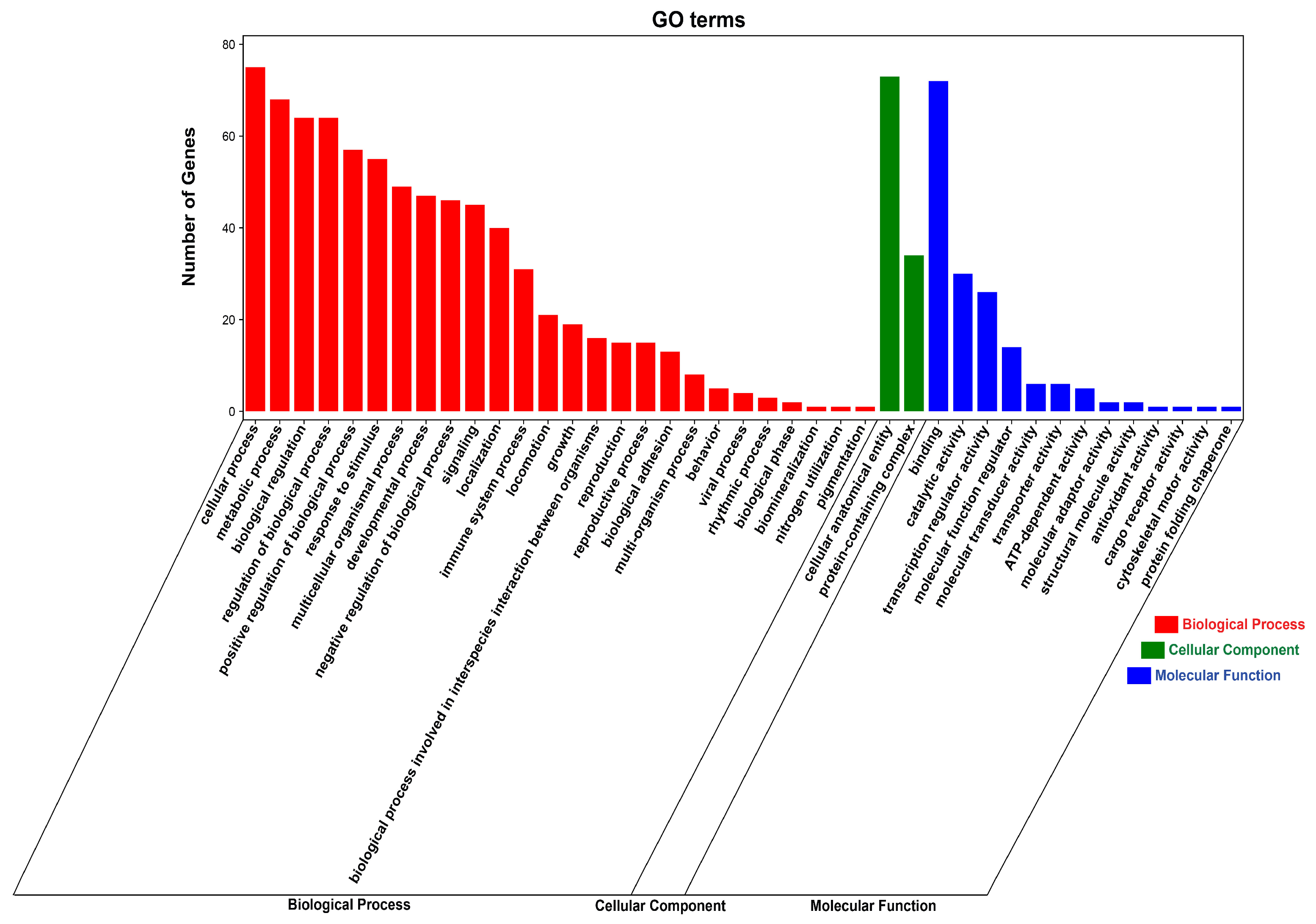
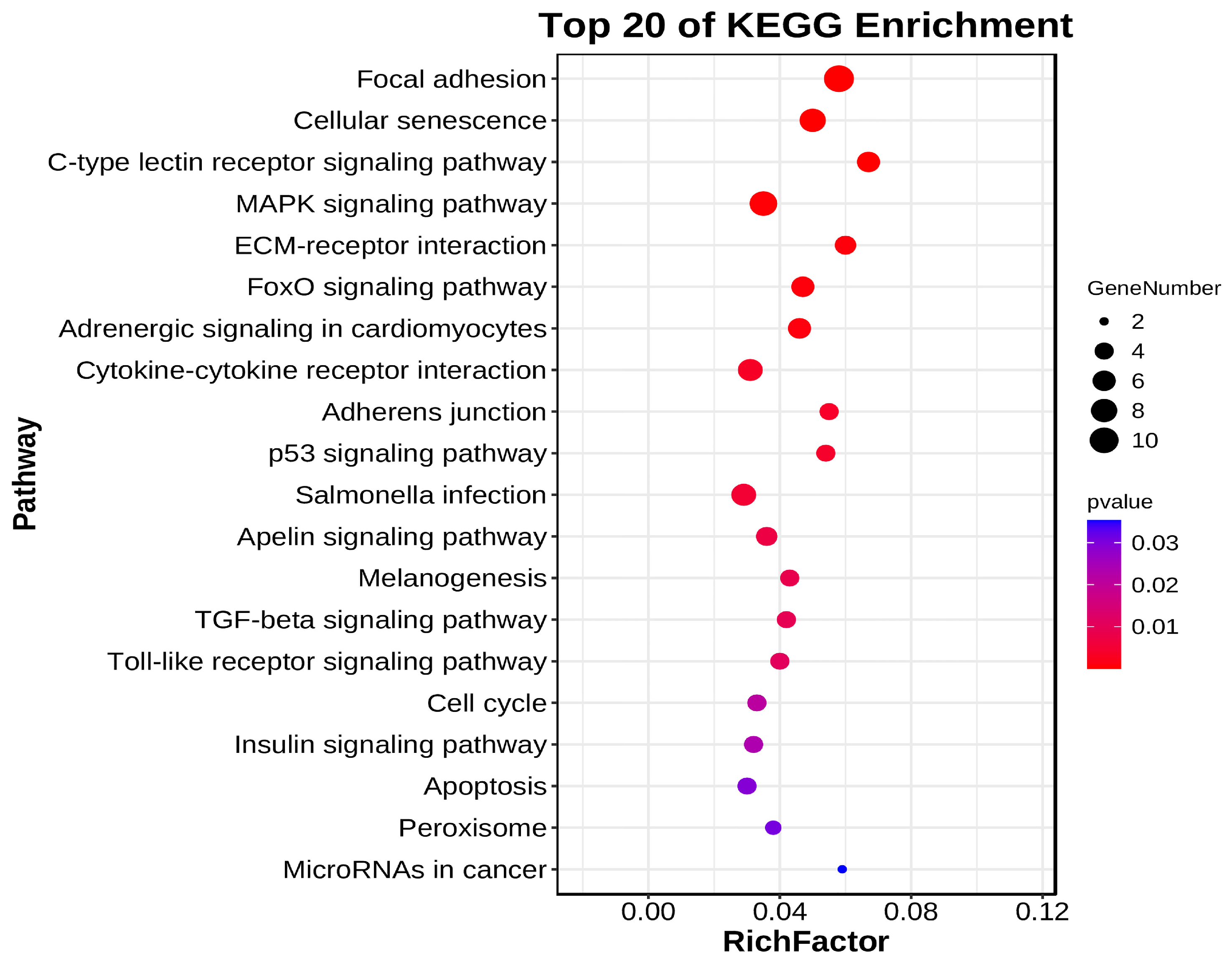
3.7. Functional Enrichment Analysis of DE circRNA Target Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Feng, M.; Zhang, X. Immunity to Avian Leukosis Virus: Where Are We Now and What Should We Do? Front. Immunol. 2016, 7, 624. [Google Scholar] [CrossRef]
- Dong, X.; Zhao, P.; Xu, B.; Fan, J.; Meng, F.; Sun, P.; Ju, S.; Li, Y.; Chang, S.; Shi, W. Avian leukosis virus in indigenous chicken breeds, China. Emerg. Microbes Infect. 2015, 4, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Lin, W.; Chang, S.; Zhao, P.; Zhang, X.; Liu, Y.; Chen, W.; Li, B.; Shu, D.; Zhang, H. Isolation, identification and evolution analysis of a novel subgroup of avian leukosis virus isolated from a local Chinese yellow broiler in South China. Arch. Virol. 2016, 161, 2717–2725. [Google Scholar] [CrossRef] [PubMed]
- Su, Q.; Li, Y.; Cui, Z.; Chang, S.; Zhao, P. The emerging novel avian leukosis virus with mutations in the gene shows competitive replication advantages both in vivo and in vitro. Emerg. Microbes Infect. 2018, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Rubin, H. The early history of tumor virology: Rous, RIF, and RAV. Proc. Natl. Acad. Sci. USA 2011, 108, 14389–14396. [Google Scholar] [CrossRef]
- Yu, M.; Zhang, Y.; Zhang, L.; Wang, S.; Liu, Y.; Xu, Z.; Liu, P.; Chen, Y.; Guo, R.; Meng, L. N123I mutation in the ALV-J receptor-binding domain region enhances viral replication ability by increasing the binding affinity with chNHE1. PLoS Pathog. 2024, 20, e1011928. [Google Scholar] [CrossRef]
- Payne, L.N.; Nair, V. The long view: 40 years of avian leukosis research. Avian Pathol. 2012, 41, 11–19. [Google Scholar] [CrossRef]
- Goyal, P.; Rajala, M.S. Reprogramming of glucose metabolism in virus infected cells. Mol. Cell. Biochem. 2023, 478, 2409–2418. [Google Scholar] [CrossRef]
- Gualdoni, G.A.; Mayer, K.A.; Kapsch, A.M.; Kreuzberg, K.; Puck, A.; Kienzl, P.; Oberndorfer, F.; Frühwirth, K.; Winkler, S.; Blaas, D. Rhinovirus induces an anabolic reprogramming in host cell metabolism essential for viral replication. Proc. Natl. Acad. Sci. USA 2018, 115, E7158–E7165. [Google Scholar] [CrossRef]
- Passalacqua, K.D.; Lu, J.; Goodfellow, I.; Kolawole, A.O.; Arche, J.R.; Maddox, R.J.; Carnahan, K.E.; O’Riordan, M.X.D.; Wobus, C.E. Glycolysis Is an Intrinsic Factor for Optimal Replication of a Norovirus. Mbio 2019, 10, e02175-18. [Google Scholar] [CrossRef]
- Fontaine, K.A.; Sanchez, E.L.; Camarda, R.; Lagunoff, M. Dengue Virus Induces and Requires Glycolysis for Optimal Replication. J. Virol. 2015, 89, 2358–2366. [Google Scholar] [CrossRef] [PubMed]
- Li, H. circRNA: A promising all-around star in the future. Epigenomics 2023, 15, 677–685. [Google Scholar] [CrossRef]
- Guo, J.; Agarwal, V.; Guo, H.; Bartel, D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014, 15, 409. [Google Scholar] [CrossRef] [PubMed]
- Lasda, E.; Parker, R. Circular RNAs: Diversity of form and function. Rna 2014, 20, 1829–1842. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna 2013, 19, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Zuo, Y.; Wang, J.; Zhang, M.; Malhotra, A.; Mayeda, A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006, 34, e63. [Google Scholar] [CrossRef] [PubMed]
- Enuka, Y.; Lauriola, M.; Feldman, M.E.; Sas-Chen, A.; Ulitsky, I.; Yarden, Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2016, 44, 1370–1383. [Google Scholar] [CrossRef]
- Rybak-Wolf, A.; Stottmeister, C.; Glazar, P.; Jens, M.; Pino, N.; Giusti, S.; Hanan, M.; Behm, M.; Bartok, O.; Ashwal-Fluss, R. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol. Cell 2015, 58, 870–885. [Google Scholar] [CrossRef]
- Ren, H.; Song, Z.; Chao, C.; Mao, W. circCCDC66 promotes thyroid cancer cell proliferation, migratory and invasive abilities and glycolysis through the miR-211-5p/PDK4 axis. Oncol. Lett. 2021, 21, 416. [Google Scholar] [CrossRef]
- Hu, Z.; Zhao, P.; Zhang, K.; Zang, L.; Liao, H.; Ma, W. Hsa_circ_0011290 regulates proliferation, apoptosis and glycolytic phenotype in papillary thyroid cancer via miR-1252/FSTL1 signal pathway. Arch. Biochem. Biophys. 2020, 685, 108353. [Google Scholar] [CrossRef]
- Shi, J.; Hu, N.; Mo, L.; Zeng, Z.; Sun, J.; Hu, Y. Deep RNA Sequencing Reveals a Repertoire of Human Fibroblast Circular RNAs Associated with Cellular Responses to Herpes Simplex Virus 1 Infection. Cell. Physiol. Biochem. 2018, 47, 2031–2045. [Google Scholar] [CrossRef] [PubMed]
- Lou, Y.; Wang, Q.; Lu, Y.; Tu, M.; Xu, X.; Xia, Y.; Peng, Y.; Lai, M.; Zheng, X. Differential circRNA expression profiles in latent human cytomegalovirus infection and validation using clinical samples. Physiol. Genom. 2019, 51, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, H.; An, M.; Zhao, B.; Ding, H.; Zhang, Z.; He, Y.; Shang, H.; Han, X. Crosstalk in competing endogenous RNA networks reveals new circular RNAs involved in the pathogenesis of early HIV infection. J. Transl. Med. 2018, 16, 332. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Qiu, L.; Chen, S.; Wang, Z.; Jiang, Y.; Bai, H.; Bi, Y.; Chen, G.; Chang, G. Circ_PIAS1 Promotes the Apoptosis of ALV-J Infected DF1 Cells by Up-Regulating miR-183. Genes 2023, 14, 1260. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef]
- Glazar, P.; Papavasileiou, P.; Rajewsky, N. circBase: A database for circular RNAs. Rna 2014, 20, 1666–1670. [Google Scholar] [CrossRef]
- Huang, H.; Lin, Y.D.; Cui, S.; Huang, Y.; Tang, Y.; Xu, J.; Bao, J.; Li, Y.; Wen, J.; Zuo, H. miRTarBase update 2022: An informative resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2022, 50, D222–D230. [Google Scholar] [CrossRef]
- Papadopoulos, G.L.; Reczko, M.; Simossis, V.A.; Sethupathy, P.; Hatzigeorgiou, A.G. The database of experimentally supported targets: A functional update of TarBase. Nucleic Acids Res. 2009, 37, D155–D158. [Google Scholar] [CrossRef]
- Carbon, S.; Douglass, E.; Good, B.M.; Unni, D.R.; Harris, N.L.; Mungall, C.J.; Basu, S.; Chisholm, R.L.; Dodson, R.J.; Hartline, E. The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Ishiguro-Watanabe, M.; Tanabe, M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2021, 49, D545–D551. [Google Scholar] [CrossRef]
- Lin, Y.; Tai, C.; Broz, V.; Tang, C.; Chen, P.; Wu, C.; Li, C.; Wu, Y. Adenosine Receptor Modulates Permissiveness of Baculovirus (Budded Virus) Infection via Regulation of Energy Metabolism in Bombyx mori. Front. Immunol. 2020, 11, 763. [Google Scholar] [CrossRef]
- Heiden, M.G.V.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Tang, N.; Liu, P.; Sun, Y.; Lu, S.; Liu, W.; Tan, L.; Song, C.; Qiu, X.; Liao, Y. Newcastle disease virus degrades SIRT3 via PINK1-PRKN-dependent mitophagy to reprogram energy metabolism in infected cells. Autophagy 2022, 18, 1503–1521. [Google Scholar] [CrossRef]
- Zhou, L.; He, R.; Fang, P.; Li, M.; Yu, H.; Wang, Q.; Yu, Y.; Wang, F.; Zhang, Y.; Chen, A. Hepatitis B virus rigs the cellular metabolome to avoid innate immune recognition. Nat. Commun. 2021, 12, 98. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Lin, C.; Qi, W.; Sun, Z.; Xie, Z.; Jia, W.; Ning, Z. Senecavirus A-induced glycolysis facilitates virus replication by promoting lactate production that attenuates the interaction between MAVS and RIG-I. PLoS Pathog. 2023, 19, e1011371. [Google Scholar] [CrossRef]
- Qiu, L.; Chang, G.; Bi, Y.; Liu, X.; Chen, G. Circular RNA and mRNA profiling reveal competing endogenous RNA networks during avian leukosis virus, subgroup J-induced tumorigenesis in chickens. PLoS ONE 2018, 13, e0204931. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, H.; Chen, X.; Wang, Z.; Yu, J.; Jia, X.; Li, Z.; Luo, W.; Abdalla, B.A.; Jebessa, E.; Nie, Q. Circular RNAs are abundant and dynamically expressed during embryonic muscle development in chickens. DNA Res. 2018, 25, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Wu, Y.; Wang, W.; Su, W.; Liu, Y.; Wang, Y.; Fan, C.; Li, X.; Li, G.; Li, Y. Circular RNAs (circRNAs) in cancer. Cancer Lett. 2018, 425, 134–142. [Google Scholar] [CrossRef]
- Tian, X.; Zhang, K.; Min, J.; Chen, C.; Cao, Y.; Ding, C.; Liu, W.; Li, J. Metabolomic Analysis of Influenza A Virus A/WSN/1933 (H1N1) Infected A549 Cells during First Cycle of Viral Replication. Viruses 2019, 11, 1007. [Google Scholar] [CrossRef]
- Ding, Z.; Ericksen, R.E.; Lee, Q.-Y.; Han, W.-P. Reprogramming of mitochondrial proline metabolism promotes liver tumorigenesis. Amino Acids 2021, 53, 1807–1815. [Google Scholar] [CrossRef]
- Yu, L.; Shi, J.; Cheng, S.; Zhu, Y.; Zhao, X.; Yang, K.; Du, X.; Klocker, H.; Yang, X.; Zhang, J. Estrogen Promotes Prostate Cancer Cell Migration via Paracrine Release of ENO1 from Stromal Cells. Mol. Endocrinol. 2012, 26, 1521–1530. [Google Scholar] [CrossRef]
- Vermeulen, N.; Arijs, I.; Joossens, S.; Vermeire, S.; Clerens, S.; van den Bergh, K.; Michiels, G.; Arckens, L.; Schuit, F.; van Lomme, L. Anti-α-enolase antibodies in patients with inflammatory bowel disease. Clin. Chem. 2008, 54, 534–541. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Gao, J.; Shen, F.; Ma, X.; Wang, Z.; Hou, X.; Hao, E.; Hou, Y.; Bai, G. Cinnamaldehyde changes the dynamic balance of glucose metabolism by targeting ENO1. Life Sci. 2020, 258, 118151. [Google Scholar] [CrossRef] [PubMed]
- Xue, Q.; Liu, H.; Zhu, Z.; Yang, F.; Song, Y.; Li, Z.; Xue, Z.; Cao, W.; Liu, X.; Zheng, H. African Swine Fever Virus Regulates Host Energy and Amino Acid Metabolism to Promote Viral Replication. J. Virol. 2022, 96, e01919-21. [Google Scholar] [CrossRef] [PubMed]
- Dryja, P.; Curtsinger, H.D.; Bartee, M.Y.; Bartee, E. Defects in intratumoral arginine metabolism attenuate the replication and therapeutic efficacy of oncolytic myxoma virus. J. Immunother. Cancer 2023, 11, e006388. [Google Scholar] [CrossRef]
- Rashid, M.U.; Lao, Y.; Spicer, V.; Coombs, K.M. Zika Virus Infection of Sertoli Cells Alters Protein Expression Involved in Activated Immune and Antiviral Response Pathways, Carbohydrate Metabolism and Cardiovascular Disease. Viruses 2022, 14, 377. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Zhu, N.; Guo, W.; Wang, X.; Li, K.; Yan, J.; Jiang, C.; Han, S.; Xiang, H.; Wu, X. RNA-Seq Revealed a Circular RNA-microRNA-mRNA Regulatory Network in Hantaan Virus Infection. Front. Cell. Infect. Microbiol. 2020, 10, 97. [Google Scholar] [CrossRef]
- Zhao, Y.; Yuan, J.; Xiao, D.; Zhang, L.; Li, C.; Hu, J.; Chen, R.; Song, D.; Wen, Y.; Wu, R. HSP90AB1 is a host factor that promotes porcine deltacoronavirus replication. J. Biol. Chem. 2024, 300, 105536. [Google Scholar] [CrossRef] [PubMed]
- Song, D.; Zhao, Y.; Sun, Y.; Liang, Y.; Chen, R.; Wen, Y.; Wu, R.; Zhao, Q.; Du, S.; Yan, Q. HSP90AB1 Is a Host Factor Required for Transmissible Gastroenteritis Virus Infection. Int. J. Mol. Sci. 2023, 24, 15971. [Google Scholar] [CrossRef] [PubMed]
- Asgari, S.; Schlapbach, L.J.; Anchisi, S.; Hammer, C.; Bartha, I.; Junier, T.; Mottet-Osman, G.; Posfay-Barbe, K.M.; Longchamp, D.; Stocker, M. Severe viral respiratory infections in children with loss-of-function mutations. Proc. Natl. Acad. Sci. USA 2017, 114, 8342–8347. [Google Scholar] [CrossRef] [PubMed]
- Tripolino, C.; Ciaffi, J.; Pucino, V.; Ruscitti, P.; van Leeuwen, N.; Borghi, C.; Giacomelli, R.; Meliconi, R.; Ursini, F. Insulin Signaling in Arthritis. Front. Immunol. 2021, 12, 672519. [Google Scholar] [CrossRef]
- Bockus, L.B.; Matsuzaki, S.; Vadvalkar, S.S.; Young, Z.T.; Giorgione, J.R.; Newhardt, M.F.; Kinter, M.; Humphries, K.M. Cardiac Insulin Signaling Regulates Glycolysis through Phosphofructokinase 2 Content and Activity. J. Am. Heart Assoc. 2017, 6, e007159. [Google Scholar] [CrossRef] [PubMed]




| Gene | Primer Sequence (5′-3′) | Product Size (bp) | Annealing Temperature (°C) |
|---|---|---|---|
| PKM2 | F: GGCACCCACGAGTATCAT | 205 | 60 |
| R: CATTGTCCAGCGTCACTTT | |||
| HK1 | F: CCTCTTGGCTTCACATTC | 231 | 60 |
| R: TTCACAGTTTGGGTCTTCAT | |||
| HIF1A | F: TTGACAAGGCATCCATTA | 157 | 60 |
| R: TCCTCAGAAAGCACCATA | |||
| LDHA | F: TGGGCATCCATCCTCTGA | 168 | 60 |
| R: CCTGCTTGTGAACCTCCT | |||
| GLUT1 | F: TGTTTGGCTTGGACTTGAT | 171 | 60 |
| R: TCTTGAGGACGCTCTTGG | |||
| PFKP | F: GCCACAACAAACCTATAACA | 241 | 60 |
| R: ATCAAAGGCAGACGAACA | |||
| β-actin | F: GAACCCCAAAGCCAACAGAGAG | 142 | 60 |
| R: ATCACCAGAGTCCATCACAATACCA |
| circRNAs | Source Gene | Primer Sequence (5′-3′) | Product Size (bp) |
|---|---|---|---|
| CircRNA116 | SPPL3 | F: TGCTTTCCTTTTGCTCCCGA | 199 |
| R: GACTGGAATCCACGAGGGAA | |||
| CircRNA429 | MICAL2 | F: ACCCCCACTAGAAATCCTTGC | 195 |
| R: CGTAGGACAGCAAATTGCCC | |||
| CircRNA436 | DENND5A | F: CCAGAGGCCAAGCACCAATG | 185 |
| R: GGCTGGCACCATCTCGG | |||
| CircRNA898 | C2CD3 | F: CAGGGACCTGCTGAGAAGAC | 166 |
| R: GATCAGCTGAGCCCATTGCC | |||
| CircRNA1056 | ENO1 | F: CCTTGTCAGAGTAGCCAGCC | 136 |
| R: CTGGGGTGTGATGGTGAGTC | |||
| CircRNA1135 | SLC9A6 | F: GAGCACGAAGGCAGAAAGCG | 194 |
| R: GCAAATGCCATGGCTCCTCG | |||
| CircRNA3765 | FAF1 | F: CCCATCGAGAAGTCCAGCGG | 155 |
| R: TGTGGGGCCATATGCTGGTC | |||
| CircRNA3869 | DOCK1 | F: GGAGTCTTTGCTGCAGCTCT | 162 |
| R: GCATCAAACACCAACGTGTCA | |||
| β-actin | / | F: GAACCCCAAAGCCAACAGAGAG | 142 |
| R: ATCACCAGAGTCCATCACAATACCA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, T.; Qiu, L.; Chen, S.; Wang, Z.; Jiang, Y.; Bai, H.; Bi, Y.; Chang, G. RNA-Seq Analysis of Glycolysis Regulation of Avian Leukosis Virus Subgroup J Replication. Animals 2024, 14, 2500. https://doi.org/10.3390/ani14172500
Yang T, Qiu L, Chen S, Wang Z, Jiang Y, Bai H, Bi Y, Chang G. RNA-Seq Analysis of Glycolysis Regulation of Avian Leukosis Virus Subgroup J Replication. Animals. 2024; 14(17):2500. https://doi.org/10.3390/ani14172500
Chicago/Turabian StyleYang, Ting, Lingling Qiu, Shihao Chen, Zhixiu Wang, Yong Jiang, Hao Bai, Yulin Bi, and Guobin Chang. 2024. "RNA-Seq Analysis of Glycolysis Regulation of Avian Leukosis Virus Subgroup J Replication" Animals 14, no. 17: 2500. https://doi.org/10.3390/ani14172500
APA StyleYang, T., Qiu, L., Chen, S., Wang, Z., Jiang, Y., Bai, H., Bi, Y., & Chang, G. (2024). RNA-Seq Analysis of Glycolysis Regulation of Avian Leukosis Virus Subgroup J Replication. Animals, 14(17), 2500. https://doi.org/10.3390/ani14172500






