Effect of Group Mixing and Available Space on Performance, Feeding Behavior, and Fecal Microbiota Composition during the Growth Period of Pigs
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design and Sample Collection
2.2. Performance and Feeding Behaviour Traits
2.3. Fecal Sampling
2.4. Microbial DNA Extraction, Sequencing and Bioinformatic Analysis
2.5. Statistical Analysis and Microbial-Biomarker Identification
3. Results
3.1. Effect of Stress on Animal Performance and Feeding Behaviour
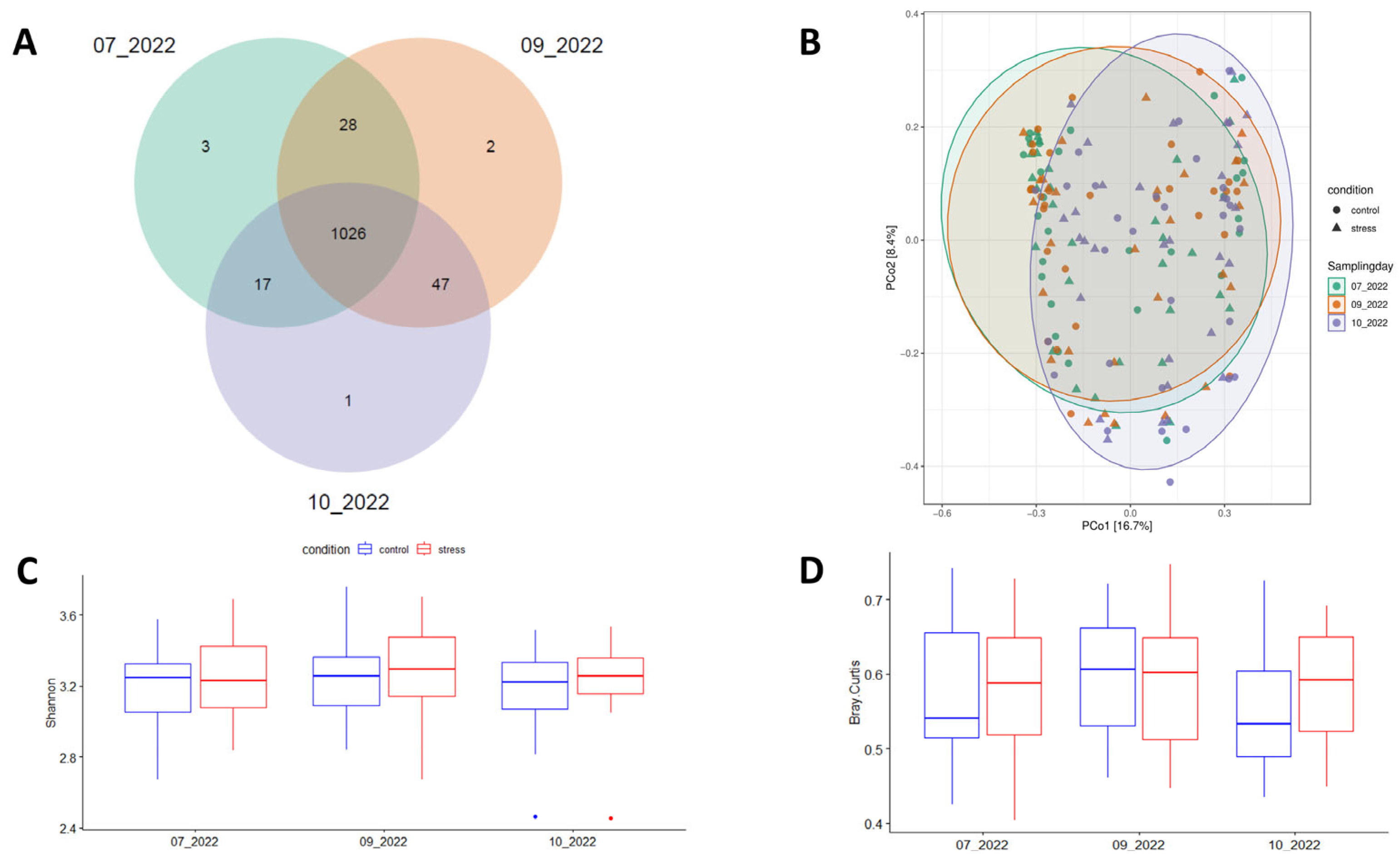
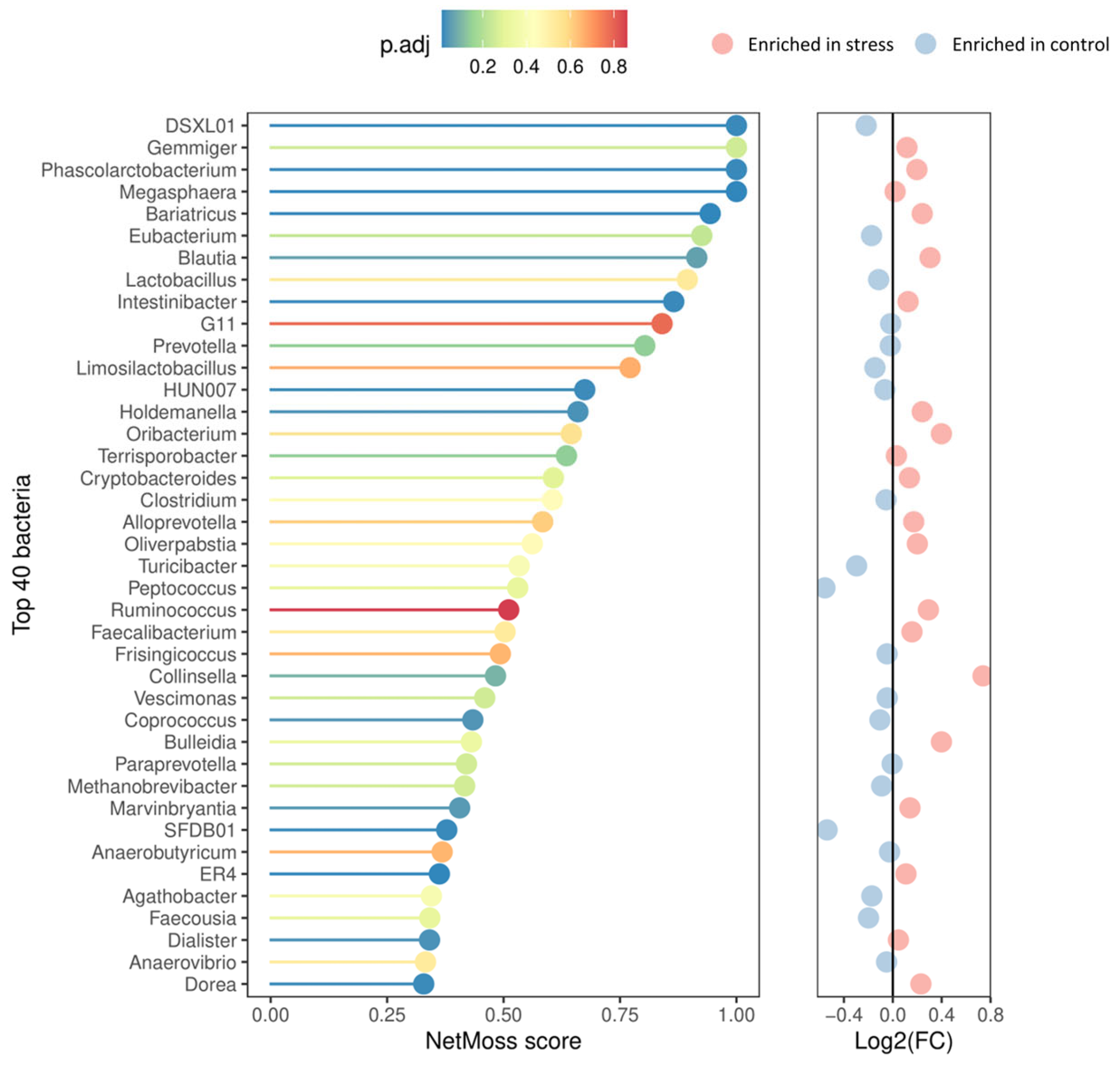
3.2. Impact of Stress on the Diversity and Composition of Fecal Microbiota
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Selye, H. The Stress of Life; Mcgraw Hill: Oxford, UK, 1978. [Google Scholar]
- Proudfoot, K.; Habing, G. Social stress as a cause of diseases in farm animals: Current knowledge and future directions. Vet. J. 2015, 206, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Litten, J.C.; Drury, P.C.; Corson, A.M.; Lean, I.J.; Clarke, L. The influence of piglet birth weight on physical and behavioural development in early life. Biol. Neonate 2003, 84, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Turner, S.P.; Ewen, M.; Rooke, J.A.; Edwards, S.A. The effect of space allowance on performance, aggression and immune competence of growing pigs housed on straw deep-litter at different group sizes. Livest. Prod. Sci. 2000, 66, 47–55. [Google Scholar] [CrossRef]
- Turner, S.P.; Roehe, R.; D’Eath, R.B.; Ison, S.H.; Farish, M.; Jack, M.C.; Lundeheim, N.; Rydhmer, L.; Lawrence, A.B. Genetic validation of postmixing skin injuries in pigs as an indicator of aggressiveness and the relationship with injuries under more stable social conditions. J. Anim. Sci. 2009, 87, 3076–3082. [Google Scholar] [CrossRef] [PubMed]
- Chidgey, K.L. Review: Space allowance for growing pigs: Animal welfare, performance and on-farm practicality. Animal 2024, 18, 100890. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, S.S.; Alvarez, J.; Bicout, D.J.; Calistri, P.; Canali, E.; Drewe, J.A.; Garin-Bastuji, B.; Gonzales Rojas, J.L.; Schmidt, G.; Herskin, M.; et al. Welfare of pigs on farm. EFSA J. 2022, 20, e07421. [Google Scholar] [CrossRef]
- Carpenter, C.B.; Holder, C.J.; Wu, F.; Woodworth, J.C.; DeRouchey, J.M.; Tokach, M.D.; Goodband, R.D.; Dritz, S.S. Effects of increasing space allowance by removing a pig or gate adjustment on finishing pig growth performance1,2. J. Anim. Sci. 2018, 96, 2659–2664. [Google Scholar] [CrossRef]
- Kim, K.H.; Kim, K.S.; Kim, J.E.; Kim, D.W.; Seol, K.H.; Lee, S.H.; Chae, B.J.; Kim, Y.H. The effect of optimal space allowance on growth performance and physiological responses of pigs at different stages of growth. Animal 2017, 11, 478–485. [Google Scholar] [CrossRef]
- Smulders, D.; Verbeke, G.; Mormède, P.; Geers, R. Validation of a behavioral observation tool to assess pig welfare. Physiol. Behav. 2006, 89, 438–447. [Google Scholar] [CrossRef]
- Kumar, B.; Manuja, A.; Aich, P. Stress and its impact on farm animals. Front. Biosci. (Elite Ed.) 2012, 4, 1759–1767. [Google Scholar] [CrossRef]
- Etim, N.; Offiong, E.; Eyoh, G.; Udo, M. Stress and Animal Welfare: An Uneasy Relationship. Eur. J. Med. Sci. 2014, 2, 9–15. [Google Scholar]
- Racewicz, P.; Ludwiczak, A.; Skrzypczak, E.; Składanowska-Baryza, J.; Biesiada, H.; Nowak, T.; Nowaczewski, S.; Zaborowicz, M.; Stanisz, M.; Ślósarz, P. Welfare Health and Productivity in Commercial Pig Herds. Animals 2021, 11, 1176. [Google Scholar] [CrossRef] [PubMed]
- de Groot, J.; Ruis, M.A.; Scholten, J.W.; Koolhaas, J.M.; Boersma, W.J. Long-term effects of social stress on antiviral immunity in pigs. Physiol. Behav. 2001, 73, 145–158. [Google Scholar] [CrossRef] [PubMed]
- Madison, A.; Kiecolt-Glaser, J.K. Stress, depression, diet, and the gut microbiota: Human–bacteria interactions at the core of psychoneuroimmunology and nutrition. Curr. Opin. Behav. Sci. 2019, 28, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Galley, J.D.; Nelson, M.C.; Yu, Z.; Dowd, S.E.; Walter, J.; Kumar, P.S.; Lyte, M.; Bailey, M.T. Exposure to a social stressor disrupts the community structure of the colonic mucosa-associated microbiota. BMC Microbiol. 2014, 14, 189. [Google Scholar] [CrossRef]
- Wu, H.-J.; Wu, E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes 2012, 3, 4–14. [Google Scholar] [CrossRef]
- Dey, P.; Saumya; Chaudhuri, R.; Raychaudhuri, S. The opportunistic nature of gut commensal microbiota The opportunistic nature of gut commensal microbiota. Crit. Rev. Microbiol. 2022, 49, 739–763. [Google Scholar] [CrossRef]
- Jiang, H.; Ling, Z.; Zhang, Y.; Mao, H.; Ma, Z.; Yin, Y.; Wang, W.; Tang, W.; Tan, Z.; Shi, J.; et al. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav. Immun. 2015, 48, 186–194. [Google Scholar] [CrossRef]
- Kiecolt-Glaser, J.K.; Wilson, S.J.; Bailey, M.L.; Andridge, R.; Peng, J.; Jaremka, L.M.; Fagundes, C.P.; Malarkey, W.B.; Laskowski, B.; Belury, M.A. Marital Distress, Depression, and a Leaky Gut: Translocation of Bacterial Endotoxin as a Pathway to Inflammation. Psychoneuroendocrinology 2018, 98, 52–60. [Google Scholar] [CrossRef]
- Tang, X.; Xiong, K.; Fang, R.; Li, M. Weaning stress and intestinal health of piglets: A review. Front. Immunol. 2022, 13, 1042778. [Google Scholar] [CrossRef]
- Hu, C.; Patil, Y.; Gong, D.; Yu, T.; Li, J.; Wu, L.; Liu, X.; Yu, Z.; Ma, X.; Yong, Y.; et al. Heat Stress-Induced Dysbiosis of Porcine Colon Microbiota Plays a Role in Intestinal Damage: A Fecal Microbiota Profile. Front. Vet. Sci. 2022, 9, 686902. [Google Scholar] [CrossRef]
- Fu, Q.; Yang, X.; Zhou, S.; Yang, Y.; Zhang, X.; Han, Q.; Ji, W.; Liu, H. Effects of short-distance transportation on physiological indexes, intestinal morphology, microbial community, and the transcriptome of the jejunum in weaned piglets. Front. Vet. Sci. 2023, 10, 1148941. [Google Scholar] [CrossRef]
- Lunney, J.K.; Van Goor, A.; Walker, K.E.; Hailstock, T.; Franklin, J.; Dai, C. Importance of the pig as a human biomedical model. Sci. Transl. Med. 2021, 13, eabd5758. [Google Scholar] [CrossRef]
- Xiao, L.; Estellé, J.; Kiilerich, P.; Ramayo-Caldas, Y.; Xia, Z.; Feng, Q.; Liang, S.; Pedersen, A.Ø.; Kjeldsen, N.J.; Liu, C.; et al. A reference gene catalogue of the pig gut microbiome. Nat. Microbiol. 2016, 1, 16161. [Google Scholar] [CrossRef]
- Dinan, T.G.; Cryan, J.F. Mood by microbe: Towards clinical translation. Genome Med. 2016, 8, 36. [Google Scholar] [CrossRef]
- Jeon, J.; Lourenco, J.; Kaiser, E.E.; Waters, E.S.; Scheulin, K.M.; Fang, X.; Kinder, H.A.; Platt, S.R.; Rothrock, M.J.; Callaway, T.R.; et al. Dynamic Changes in the Gut Microbiome at the Acute Stage of Ischemic Stroke in a Pig Model. Front. Neurosci. 2020, 14, 587986. [Google Scholar] [CrossRef]
- Nguyen, T.Q.; Martínez-Álvaro, M.; Lima, J.; Auffret, M.D.; Rutherford, K.M.D.; Simm, G.; Dewhurst, R.J.; Baima, E.T.; Roehe, R. Identification of intestinal and fecal microbial biomarkers using a porcine social stress model. Front. Microbiol. 2023, 14, 1197371. [Google Scholar] [CrossRef]
- da Fonseca de Oliveira, A.C.; Costa, L.B.; Weber, S.H.; Ramayo-Caldas, Y.; Dalmau, A. Mixed management in growing and finishing pigs: Differences between gender and their impacts on behavior, growth performance, and physiological parameters. PLoS ONE 2023, 18, e0284481. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- McDonald, D.; Jiang, Y.; Balaban, M.; Cantrell, K.; Zhu, Q.; Gonzalez, A.; Morton, J.T.; Nicolaou, G.; Parks, D.H.; Karst, S.M.; et al. Greengenes2 unifies microbial data in a single reference tree. Nat. Biotechnol. 2023, 42, 715–718. [Google Scholar] [CrossRef]
- Liu, C.; Cui, Y.; Li, X.; Yao, M. microeco: An R package for data mining in microbial community ecology. FEMS Microbiol. Ecol. 2021, 97, fiaa255. [Google Scholar] [CrossRef]
- Shannon, C.E. A Mathematical Theory of Communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef]
- Whittaker, R.H. Evolution and Measurement of Species Diversity. TAXON 1972, 21, 213–251. [Google Scholar] [CrossRef]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001, 26, 32–46. [Google Scholar] [CrossRef]
- Xiao, L.; Zhang, F.; Zhao, F. Large-scale microbiome data integration enables robust biomarker identification. Nat. Comput. Sci. 2022, 2, 307–316. [Google Scholar] [CrossRef]
- Friedman, J.; Alm, E.J. Inferring Correlation Networks from Genomic Survey Data. PLoS Comput. Biol. 2012, 8, e1002687. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Scardoni, G.; Tosadori, G.; Faizan, M.; Spoto, F.; Fabbri, F.; Laudanna, C. Biological Network Analysis with CentiScaPe: Centralities and Experimental Dataset Integration. F1000Research 2015. Available online: https://f1000research.com/articles/3-139 (accessed on 26 April 2024).
- Chen, C.; Zhou, Y.; Fu, H.; Xiong, X.; Fang, S.; Jiang, H.; Wu, J.; Yang, H.; Gao, J.; Huang, L. Expanded catalog of microbial genes and metagenome-assembled genomes from the pig gut microbiome. Nat. Commun. 2021, 12, 1106. [Google Scholar] [CrossRef]
- Luo, L.; van der Zande, L.E.; van Marwijk, M.A.; Knol, E.F.; Rodenburg, T.B.; Bolhuis, J.E.; Parois, S.P. Impact of Enrichment and Repeated Mixing on Resilience in Pigs. Front. Vet. Sci. 2022, 9, 829060. [Google Scholar] [CrossRef]
- Moore, A.S.; Gonyou, H.W.; Stookey, J.M.; McLaren, D.G. Effect of group composition and pen size on behavior, productivity and immune response of growing pigs. Appl. Anim. Behav. Sci. 1994, 40, 13–30. [Google Scholar] [CrossRef]
- Tuchscherer, M.; Puppe, B.; Tuchscherer, A.; Kanitz, E. Effects of social status after mixing on immune, metabolic, and endocrine responses in pigs. Physiol. Behav. 1998, 64, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Camp Montoro, J.; Pessoa, J.; Solà-Oriol, D.; Muns, R.; Gasa, J.; Manzanilla, E.G. Effect of Phase Feeding, Space Allowance and Mixing on Productive Performance of Grower-Finisher Pigs. Animals 2022, 12, 390. [Google Scholar] [CrossRef] [PubMed]
- Hemsworth, P.H.; Morrison, R.S.; Tilbrook, A.J.; Butler, K.L.; Rice, M.; Moeller, S.J. Effects of varying floor space on aggressive behavior and cortisol concentrations in group-housed sows1. J. Anim. Sci. 2016, 94, 4809–4818. [Google Scholar] [CrossRef]
- Barnett, J.L.; Hemsworth, P.H.; Cronin, G.M.; Newman, E.A.; McCallum, T.H.; Chilton, D. Effects of pen size, partial stalls and method of feeding on welfare-related behavioural and physiological responses of group-housed pigs. Appl. Anim. Behav. Sci. 1992, 34, 207–220. [Google Scholar] [CrossRef]
- Salak-Johnson, J.L.; Niekamp, S.R.; Rodriguez-Zas, S.L.; Ellis, M.; Curtis, S.E. Space allowance for dry, pregnant sows in pens: Body condition, skin lesions, and performance. J. Anim. Sci. 2007, 85, 1758–1769. [Google Scholar] [CrossRef]
- da Fonseca de Oliveira, A.C.; Webber, S.H.; Ramayo-Caldas, Y.; Dalmau, A.; Costa, L.B. Hierarchy Establishment in Growing Finishing Pigs: Impacts on Behavior, Growth Performance, and Physiological Parameters. Animals 2023, 13, 292. [Google Scholar] [CrossRef]
- Hyun, Y.; Ellis, M.; Johnson, R.W. Effects of feeder type, space allowance, and mixing on the growth performance and feed intake pattern of growing pigs. J. Anim. Sci. 1998, 76, 2771–2778. [Google Scholar] [CrossRef]
- Li, Y.; Song, Z.; Kerr, K.A.; Moeser, A.J. Chronic social stress in pigs impairs intestinal barrier and nutrient transporter function, and alters neuro-immune mediator and receptor expression. PLoS ONE 2017, 12, e0171617. [Google Scholar] [CrossRef]
- Martínez-Miró, S.; Tecles, F.; Ramón, M.; Escribano, D.; Hernández, F.; Madrid, J.; Orengo, J.; Martínez-Subiela, S.; Manteca, X.; Cerón, J.J. Causes, consequences and biomarkers of stress in swine: An update. BMC Vet. Res. 2016, 12, 171. [Google Scholar] [CrossRef]
- Knight, R.; Cedillo, Y.; Judd, S.; Baker, E.; Fruge, A.; Moellering, D. A cross-sectional study observing the association of psychosocial stress and dietary intake with gut microbiota genera and alpha diversity among a young adult cohort of black and white women in Birmingham, Alabama. BMC Women’s Health 2023, 24, 142. [Google Scholar]
- Bailey, M.T.; Dowd, S.E.; Galley, J.D.; Hufnagle, A.R.; Allen, R.G.; Lyte, M. Exposure to a social stressor alters the structure of the intestinal microbiota: Implications for stressor-induced immunomodulation. Brain Behav. Immun. 2011, 25, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Cheung, S.G.; Goldenthal, A.R.; Uhlemann, A.-C.; Mann, J.J.; Miller, J.M.; Sublette, M.E. Systematic Review of Gut Microbiota and Major Depression. Front. Psychiatry 2019, 10, 34. [Google Scholar] [CrossRef]
- Simpson, C.A.; Diaz-Arteche, C.; Eliby, D.; Schwartz, O.S.; Simmons, J.G.; Cowan, C.S.M. The gut microbiota in anxiety and depression—A systematic review. Clin. Psychol. Rev. 2021, 83, 101943. [Google Scholar] [CrossRef] [PubMed]
- Kubasova, T.; Davidova-Gerzova, L.; Babak, V.; Cejkova, D.; Montagne, L.; Le-Floc’h, N.; Rychlik, I. Effects of host genetics and environmental conditions on fecal microbiota composition of pigs. PLoS ONE 2018, 13, e0201901. [Google Scholar] [CrossRef]
- McCormack, U.M.; Curião, T.; Buzoianu, S.G.; Prieto, M.L.; Ryan, T.; Varley, P.; Crispie, F.; Magowan, E.; Metzler-Zebeli, B.U.; Berry, D.; et al. Exploring a Possible Link between the Intestinal Microbiota and Feed Efficiency in Pigs. Appl. Environ. Microbiol. 2017, 83, e00380-17. [Google Scholar] [CrossRef]
- Tourlousse, D.M.; Sakamoto, M.; Miura, T.; Narita, K.; Ohashi, A.; Uchino, Y.; Yamazoe, A.; Kameyama, K.; Terauchi, J.; Ohkuma, M.; et al. Complete Genome Sequence of Collinsella aerofaciens JCM 10188T. Microbiol. Resour. Announc. 2020, 9, e00134-20. [Google Scholar] [CrossRef]
- Strati, F.; Cavalieri, D.; Albanese, D.; De Felice, C.; Donati, C.; Hayek, J.; Jousson, O.; Leoncini, S.; Renzi, D.; Calabrò, A.; et al. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome 2017, 5, 24. [Google Scholar] [CrossRef]
- Astbury, S.; Atallah, E.; Vijay, A.; Aithal, G.P.; Grove, J.I.; Valdes, A.M. Lower gut microbiome diversity and higher abundance of proinflammatory genus Collinsella are associated with biopsy-proven nonalcoholic steatohepatitis. Gut Microbes 2020, 11, 569–580. [Google Scholar] [CrossRef]
- Geng, S.; Yang, L.; Cheng, F.; Zhang, Z.; Li, J.; Liu, W.; Li, Y.; Chen, Y.; Bao, Y.; Chen, L.; et al. Gut Microbiota Are Associated With Psychological Stress-Induced Defections in Intestinal and Blood–Brain Barriers. Front. Microbiol. 2020, 10, 3067. [Google Scholar] [CrossRef]
- Ochoteco-Asensio, J.; Zigovski, G.; Batista Costa, L.; Rio-López, R.; Clavell-Sansalvador, A.; Ramayo-Caldas, Y.; Dalmau, A. Effect on Feeding Behaviour and Growing of Being a Dominant or Subordinate Growing Pig and Its Relationship with the Faecal Microbiota. Animals 2024, 14, 1906. [Google Scholar] [CrossRef] [PubMed]
- Wanapaisan, P.; Chuansangeam, M.; Nopnipa, S.; Mathuranyanon, R.; Nonthabenjawan, N.; Ngamsombat, C.; Thientunyakit, T.; Muangpaisan, W. Association between Gut Microbiota with Mild Cognitive Impairment and Alzheimer’s Disease in a Thai Population. Neurodegener. Dis. 2022, 22, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Scheepers, I.M.; Cryan, J.F.; Bastiaanssen, T.F.S.; Rea, K.; Clarke, G.; Jaspan, H.B.; Harvey, B.H.; Hemmings, S.M.J.; Santana, L.; van der Sluis, R.; et al. Natural compulsive-like behaviour in the deer mouse (Peromyscus maniculatus bairdii) is associated with altered gut microbiota composition. Eur. J. Neurosci. 2020, 51, 1419–1427. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Mao, B.; Gu, J.; Wu, J.; Cui, S.; Wang, G.; Zhao, J.; Zhang, H.; Chen, W. Blautia-a new functional genus with potential probiotic properties? Gut Microbes 2021, 13, 1875796. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Yan, Y.; Webb, R.J.; Li, Y.; Mehrabani, S.; Xin, B.; Sun, X.; Wang, Y.; Mazidi, M. Psychological Stress and Gut Microbiota Composition: A Systematic Review of Human Studies. Neuropsychobiology 2023, 82, 247–262. [Google Scholar] [CrossRef] [PubMed]
- Michels, N.; Van de Wiele, T.; Fouhy, F.; O’Mahony, S.; Clarke, G.; Keane, J. Gut microbiome patterns depending on children’s psychosocial stress: Reports versus biomarkers. Brain Behav. Immun. 2019, 80, 751–762. [Google Scholar] [CrossRef]
- He, H.; Zhao, Z.; Xiao, C.; Li, L.; Liu, Y.; Fu, J.; Liao, H.; Zhou, T.; Zhang, J. Gut microbiome promotes mice recovery from stress-induced depression by rescuing hippocampal neurogenesis. Neurobiol. Dis. 2024, 191, 106396. [Google Scholar] [CrossRef]
- Valles-Colomer, M.; Falony, G.; Darzi, Y.; Tigchelaar, E.F.; Wang, J.; Tito, R.Y.; Schiweck, C.; Kurilshikov, A.; Joossens, M.; Wijmenga, C.; et al. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat. Microbiol. 2019, 4, 623–632. [Google Scholar] [CrossRef]
- Knudsen, J.K.; Michaelsen, T.Y.; Bundgaard-Nielsen, C.; Nielsen, R.E.; Hjerrild, S.; Leutscher, P.; Wegener, G.; Sørensen, S. Faecal microbiota transplantation from patients with depression or healthy individuals into rats modulates mood-related behaviour. Sci. Rep. 2021, 11, 21869. [Google Scholar] [CrossRef]
- McGuinness, A.J.; Davis, J.A.; Dawson, S.L.; Loughman, A.; Collier, F.; O’Hely, M.; Simpson, C.A.; Green, J.; Marx, W.; Hair, C.; et al. A systematic review of gut microbiota composition in observational studies of major depressive disorder, bipolar disorder and schizophrenia. Mol. Psychiatry 2022, 27, 1920–1935. [Google Scholar] [CrossRef]
- Li, S.; Song, J.; Ke, P.; Kong, L.; Lei, B.; Zhou, J.; Huang, Y.; Li, H.; Li, G.; Chen, J.; et al. The gut microbiome is associated with brain structure and function in schizophrenia. Sci. Rep. 2021, 11, 9743. [Google Scholar] [CrossRef]
- Louis, P.; Flint, H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 2017, 19, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Vourlaki, I.-T.; Rio, R.; Clavell, A.; Ramírez-Ayala, L.C.; Ballester, M.; Sanchez, J.P.; Piles, M.; Quintanilla, R.; de Oliveira, A.C.F.; Costa, L.B.; et al. Enterosignatures of the Fecal Pig Microbiota: Exploring Determinants and Revealing Host-Performance Consequences. 2024. Available online: https://www.researchsquare.com/article/rs-3978889/v1 (accessed on 19 July 2024). [CrossRef]
- Kullberg, R.F.J.; Wikki, I.; Haak, B.W.; Kauko, A.; Galenkamp, H.; Peters-Sengers, H.; Butler, J.M.; Havulinna, A.S.; Palmu, J.; McDonald, D.; et al. Association between butyrate-producing gut bacteria and the risk of infectious disease hospitalisation: Results from two observational, population-based microbiome studies. Lancet Microbe 2024, 5, 100864. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Yang, Q.; Mi, X.; Qiu, L.; Tao, X.; Zhang, Z.; Xia, J.; Wu, Q.; Wei, H. Ripened Pu-erh Tea Extract Promotes Gut Microbiota Resilience against Dextran Sulfate Sodium Induced Colitis. J. Agric. Food Chem. 2021, 69, 2190–2203. [Google Scholar] [CrossRef]
- van de Wouw, M.; Boehme, M.; Lyte, J.M.; Wiley, N.; Strain, C.; O’Sullivan, O.; Clarke, G.; Stanton, C.; Dinan, T.G.; Cryan, J.F. Short-chain fatty acids: Microbial metabolites that alleviate stress-induced brain–gut axis alterations. J. Physiol. 2018, 596, 4923–4944. [Google Scholar] [CrossRef]
- Chang, L.; Wei, Y.; Hashimoto, K. Brain–gut–microbiota axis in depression: A historical overview and future directions. Brain Res. Bull. 2022, 182, 44–56. [Google Scholar] [CrossRef]


| Trait | Control (se) | Stress (se) | p-Value |
|---|---|---|---|
| Body weight (BW kg) | 136 (1.49) | 127 (1.35) | <0.0001 |
| Average daily gain (ADG kg) | 1.01 (0.01) | 0.95 (0.01) | 0.0001 |
| Feed conversion ratio (FCR kg) | 2.93 (0.18) | 2.87 (0.16) | 0.048 |
| Total number of visits | 719 (0.75) | 589 (0.54) | <2 × 10−16 |
| Total feed time per day (min) | 74.55 (3.61) | 75.83 (3.97) | 0.028 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clavell-Sansalvador, A.; Río-López, R.; González-Rodríguez, O.; García-Gil, L.J.; Xifró, X.; Zigovski, G.; Ochoteco-Asensio, J.; Ballester, M.; Dalmau, A.; Ramayo-Caldas, Y. Effect of Group Mixing and Available Space on Performance, Feeding Behavior, and Fecal Microbiota Composition during the Growth Period of Pigs. Animals 2024, 14, 2704. https://doi.org/10.3390/ani14182704
Clavell-Sansalvador A, Río-López R, González-Rodríguez O, García-Gil LJ, Xifró X, Zigovski G, Ochoteco-Asensio J, Ballester M, Dalmau A, Ramayo-Caldas Y. Effect of Group Mixing and Available Space on Performance, Feeding Behavior, and Fecal Microbiota Composition during the Growth Period of Pigs. Animals. 2024; 14(18):2704. https://doi.org/10.3390/ani14182704
Chicago/Turabian StyleClavell-Sansalvador, Adrià, Raquel Río-López, Olga González-Rodríguez, L. Jesús García-Gil, Xavier Xifró, Gustavo Zigovski, Juan Ochoteco-Asensio, Maria Ballester, Antoni Dalmau, and Yuliaxis Ramayo-Caldas. 2024. "Effect of Group Mixing and Available Space on Performance, Feeding Behavior, and Fecal Microbiota Composition during the Growth Period of Pigs" Animals 14, no. 18: 2704. https://doi.org/10.3390/ani14182704
APA StyleClavell-Sansalvador, A., Río-López, R., González-Rodríguez, O., García-Gil, L. J., Xifró, X., Zigovski, G., Ochoteco-Asensio, J., Ballester, M., Dalmau, A., & Ramayo-Caldas, Y. (2024). Effect of Group Mixing and Available Space on Performance, Feeding Behavior, and Fecal Microbiota Composition during the Growth Period of Pigs. Animals, 14(18), 2704. https://doi.org/10.3390/ani14182704







