Detection and Localization of IL-8 and CXCR1 in Rainbow Trout Larvae in Response to Pseudomonas aeruginosa Lipopolysaccharide
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
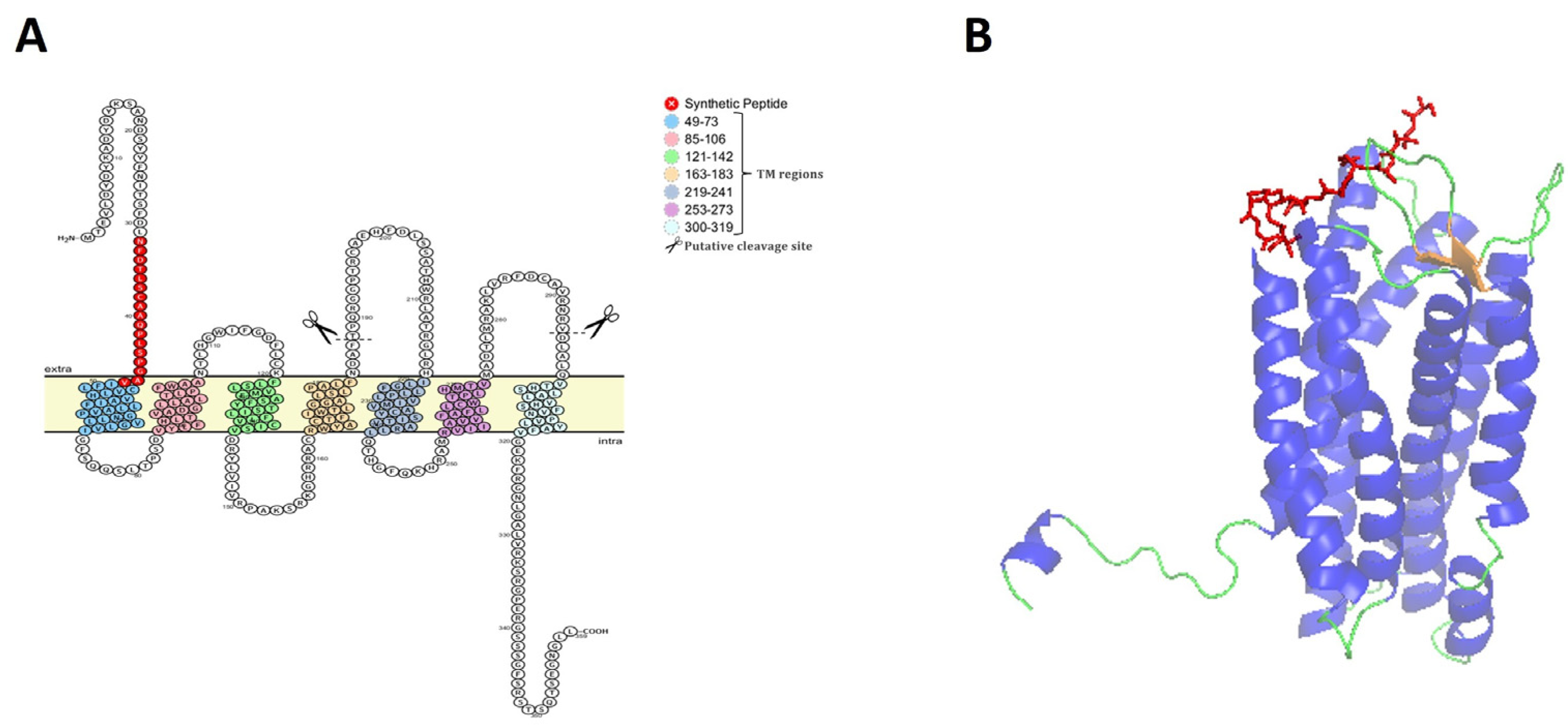
2.1. In Silico Analysis, Synthesis, Characterization, and Antisera Production
2.2. Obtaining Trout Larvae and Challenge with LPS
2.3. Immunodetection of omCXCR1 in Trout Larvae
2.4. Immunohistochemistry and Immunofluorescence
2.5. Statistical Analysis
2.6. Ethics Statement
3. Results
3.1. Characterization of CXCR1 of Rainbow Trout and Selection of Epitope Peptide
3.2. Characterization of CXCR1 Mice Antisera
3.3. Effect of LPS Challengue in the Protein Expression and Location of omCXCR1 and omIL-8 in Trout Larvae
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- D’Agaro, E.; Gibertoni, P.P.; Esposito, S. Recent Trends and Economic Aspects in the Rainbow Trout (Oncorhynchus mykiss) Sector. Appl. Sci. 2022, 12, 8773. [Google Scholar] [CrossRef]
- Criddle, K.R.; Shimizu, I. Economic Importance of Wild Salmon; Nova Science Publishers, Inc.: Hauppauge, NY, USA, 2014; ISBN 9781631175701. [Google Scholar]
- Vadstein, O.; Bergh, Ø.; Gatesoupe, F.-J.; Galindo-Villegas, J.; Mulero, V.; Picchietti, S.; Scapigliati, G.; Makridis, P.; Olsen, Y.; Dierckens, K.; et al. Microbiology and Immunology of Fish Larvae. Rev. Aquac. 2013, 5, S1–S25. [Google Scholar] [CrossRef]
- Clark, E.S.; Pompini, M.; Marques da Cunha, L.; Wedekind, C. Maternal and Paternal Contributions to Pathogen Resistance Dependent on Development Stage in a Whitefish (Salmonidae). Funct. Ecol. 2014, 28, 714–723. [Google Scholar] [CrossRef]
- Nayak, S.P.; Mohanty, B.R.; Mishra, J.; Rauta, P.R.; Das, A.; Eknath, A.E.; Sahoo, P.K. Ontogeny and Tissue-Specific Expression of Innate Immune Related Genes in Rohu, Labeo Rohita (Hamilton). Fish Shellfish Immunol. 2011, 30, 1197–1201. [Google Scholar] [CrossRef] [PubMed]
- Rauta, P.R.; Nayak, B.; Das, S. Immune System and Immune Responses in Fish and Their Role in Comparative Immunity Study: A Model for Higher Organisms. Immunol. Lett. 2012, 148, 23–33. [Google Scholar] [CrossRef]
- Seppola, M.; Johnsen, H.; Mennen, S.; Myrnes, B.; Tveiten, H. Maternal Transfer and Transcriptional Onset of Immune Genes during Ontogenesis in Atlantic Cod. Dev. Comp. Immunol. 2009, 33, 1205–1211. [Google Scholar] [CrossRef]
- Heinecke, R.D.; Buchmann, K. Inflammatory Response of Rainbow Trout Oncorhynchus mykiss (Walbaum, 1792) Larvae against Ichthyophthirius Multifiliis. Fish Shellfish Immunol. 2013, 34, 521–528. [Google Scholar] [CrossRef]
- Chettri, J.K.; Raida, M.K.; Kania, P.W.; Buchmann, K. Differential Immune Response of Rainbow Trout (Oncorhynchus mykiss) at Early Developmental Stages (Larvae and Fry) against the Bacterial Pathogen Yersinia Ruckeri. Dev. Comp. Immunol. 2012, 36, 463–474. [Google Scholar] [CrossRef]
- Oh, W.T.; Kim, J.H.; Jun, J.W.; Giri, S.S.; Yun, S.; Kim, H.J.; Kim, S.G.; Kim, S.W.; Han, S.J.; Kwon, J.; et al. Genetic Characterization and Pathological Analysis of a Novel Bacterial Pathogen, Pseudomonas Tructae, in Rainbow Trout (Oncorhynchus mykiss). Microorganisms 2019, 7, 432. [Google Scholar] [CrossRef]
- Oberlé, K.; Bouju-Albert, A.; Helsens, N.; Pangga, G.; Prevost, H.; Magras, C.; Calvez, S. No Evidence for a Relationship between Farm or Transformation Process Locations and Antibiotic Resistance Patterns of Pseudomonas Population Associated with Rainbow Trout (Oncorhynchus mykiss). J. Appl. Microbiol. 2022, 132, 1738–1750. [Google Scholar] [CrossRef]
- Algammal, A.M.; Mabrok, M.; Sivaramasamy, E.; Youssef, F.M.; Atwa, M.H.; El-kholy, A.W.; Hetta, H.F.; Hozzein, W.N. Emerging MDR-Pseudomonas Aeruginosa in Fish Commonly Harbor OprL and ToxA Virulence Genes and BlaTEM, BlaCTX-M, and TetA Antibiotic-Resistance Genes. Sci. Rep. 2020, 10, 15961. [Google Scholar] [CrossRef] [PubMed]
- Santana, P.A.; Guzmán, F.; Forero, J.C.; Luna, O.F.; Mercado, L. Hepcidin, Cathelicidin-1 and IL-8 as Immunological Markers of Responsiveness in Early Developmental Stages of Rainbow Trout. Dev. Comp. Immunol. 2016, 62, 48–57. [Google Scholar] [CrossRef]
- Chen, J.; Xu, Q.; Wang, T.; Collet, B.; Corripio-Miyar, Y.; Bird, S.; Xie, P.; Nie, P.; Secombes, C.J.; Zou, J. Phylogenetic Analysis of Vertebrate CXC Chemokines Reveals Novel Lineage Specific Groups in Teleost Fish. Dev. Comp. Immunol. 2013, 41, 137–152. [Google Scholar] [CrossRef] [PubMed]
- Alejo, A.; Tafalla, C. Chemokines in Teleost Fish Species. Dev. Comp. Immunol. 2011, 35, 1215–1222. [Google Scholar] [CrossRef] [PubMed]
- van der Aa, L.M.; Chadzinska, M.; Tijhaar, E.; Boudinot, P.; Lidy verburg-Van kemenade, B.M. CXCL8 Chemokines in Teleost Fish: Two Lineages with Distinct Expression Profiles during Early Phases of Inflammation. PLoS ONE 2010, 5, e12384. [Google Scholar] [CrossRef] [PubMed]
- Coombs, C.; Georgantzoglou, A.; Walker, H.A.; Patt, J.; Merten, N.; Poplimont, H.; Busch-Nentwich, E.M.; Williams, S.; Kotsi, C.; Kostenis, E.; et al. Chemokine Receptor Trafficking Coordinates Neutrophil Clustering and Dispersal at Wounds in Zebrafish. Nat. Commun. 2019, 10, 5166. [Google Scholar] [CrossRef]
- Kolaczkowska, E.; Kubes, P. Neutrophil Recruitment and Function in Health and Inflammation. Nat. Rev. Immunol. 2013, 13, 159–175. [Google Scholar] [CrossRef]
- Li, H.; Cai, Y.; Xie, P.; Li, G.; Hao, L.; Xiong, Q. Identification and Expression Profiles of IL-8 in Bighead Carp (Aristichthys nobilis) in Response to Microcystin-LR. Arch. Environ. Contam. Toxicol. 2013, 65, 537–545. [Google Scholar] [CrossRef]
- Cotton, M.; Claing, A. G Protein-Coupled Receptors Stimulation and the Control of Cell Migration. Cell. Signal. 2009, 21, 1045–1053. [Google Scholar] [CrossRef]
- Xu, Q.; Li, R.; Monte, M.M.; Jiang, Y.; Nie, P.; Holland, J.W.; Secombes, C.J.; Wang, T. Sequence and Expression Analysis of Rainbow Trout CXCR2, CXCR3a and CXCR3b Aids Interpretation of Lineage-Specific Conversion, Loss and Expansion of These Receptors during Vertebrate Evolution. Dev. Comp. Immunol. 2014, 45, 201–213. [Google Scholar] [CrossRef]
- Zlotnik, A.; Yoshie, O. The Chemokine Superfamily Revisited NIH Public Access. Immunity 2012, 36, 705–716. [Google Scholar] [CrossRef] [PubMed]
- Bricknell, I.; Dalmo, R.A. The Use of Immunostimulants in Fish Larval Aquaculture. Fish Shellfish Immunol. 2005, 19, 457–472. [Google Scholar] [CrossRef] [PubMed]
- Santana, P.; Palacios, C.; Narváez, E.; Guzmán, F.; Gallardo, J.A.; Mercado, L. Anti-Peptide Antibodies: A Tool for Detecting IL-8 in Salmonids. Electron. J. Biotechnol. 2012, 15, 20. [Google Scholar] [CrossRef]
- Omasits, U.; Ahrens, C.H.; Müller, S.; Wollscheid, B. Protter: Interactive Protein Feature Visualization and Integration with Experimental Proteomic Data. Bioinformatics 2014, 30, 884–886. [Google Scholar] [CrossRef] [PubMed]
- Santana, P.A.; Álvarez, C.A.; Guzmán, F.; Mercado, L. Fish & Shellfish Immunology Development of a Sandwich ELISA for Quantifying Hepcidin in Rainbow Trout. Fish Shellfish Immunol. 2013, 35, 748–755. [Google Scholar] [PubMed]
- Bethke, J.; Rojas, V.; Berendsen, J.; Cárdenas, C.; Guzmán, F.; Gallardo, J.A.; Mercado, L. Development of a New Antibody for Detecting Natural Killer Enhancing Factor (NKEF)-like Protein in Infected Salmonids. J. Fish Dis. 2012, 35, 379–388. [Google Scholar] [CrossRef]
- Tedesco, P.; Saraiva, M.; Sandoval-Sierra, J.V.; Fioravanti, M.L.; Morandi, B.; Dieguez-Uribeondo, J.; Van West, P.; Galuppi, R. Evaluation of Potential Transfer of the Pathogen Saprolegnia Parasitica between Farmed Salmonids and Wild Fish. Pathogens 2021, 10, 926. [Google Scholar] [CrossRef]
- Fayaz, I.; Bhat, R.A.H.; Tandel, R.S.; Dash, P.; Chandra, S.; Dubey, M.K.; Ganie, P.A. Comprehensive Review on Infectious Pancreatic Necrosis Virus. Aquaculture 2023, 574, 739737. [Google Scholar] [CrossRef]
- Novoa, B.; Bowman, T.V.; Zon, L.; Figueras, A. LPS Response and Tolerance in the Zebrafish (Danio rerio). Fish Shellfish Immunol. 2009, 26, 326–331. [Google Scholar] [CrossRef]
- Santos, R.A.; Cardoso, C.; Pedrosa, N.; Gonçalves, G.; Matinha-Cardoso, J.; Coutinho, F.; Carvalho, A.P.; Tamagnini, P.; Oliva-Teles, A.; Oliveira, P.; et al. LPS-Induced Mortality in Zebrafish: Preliminary Characterisation of Common Fish Pathogens. Microorganisms 2023, 11, 2205. [Google Scholar] [CrossRef]
- Biedroń, R.; Peruń, A.; Józefowski, S. CD36 Differently Regulates Macrophage Responses to Smooth and Rough Lipopolysaccharide. PLoS ONE 2016, 11, e0153558. [Google Scholar] [CrossRef] [PubMed]
- Giordano, N.P.; Cian, M.B.; Dalebroux, Z.D. Outer Membrane Lipid Secretion and the Innate Immune Response to Gram-Negative Bacteria. Infect. Immun. 2020, 88, e00920-19. [Google Scholar] [CrossRef] [PubMed]
- Soliman, A.M.; Barreda, D.R. The Acute Inflammatory Response of Teleost Fish. Dev. Comp. Immunol. 2023, 146, 104731. [Google Scholar] [CrossRef] [PubMed]
- Sepulcre, M.P.; Alcaraz-Pérez, F.; López-Muñoz, A.; Roca, F.J.; Meseguer, J.; Cayuela, M.L.; Mulero, V. Evolution of Lipopolysaccharide (LPS) Recognition and Signaling: Fish TLR4 Does Not Recognize LPS and Negatively Regulates NF-ΚB Activation. J. Immunol. 2009, 182, 1836–1845. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xia, P.; Wu, J.; Huang, A.; Bu, G.; Meng, F.; Kong, F.; Cao, X.; Han, X.; Yu, G.; et al. The Potential Sensing Molecules and Signal Cascades for Protecting Teleost Fishes against Lipopolysaccharide. Fish Shellfish Immunol. 2020, 97, 235–247. [Google Scholar] [CrossRef]
- MacKenzie, S.A.; Roher, N.; Boltaña, S.; Goetz, F.W. Peptidoglycan, Not Endotoxin, Is the Key Mediator of Cytokine Gene Expression Induced in Rainbow Trout Macrophages by Crude LPS. Mol. Immunol. 2010, 47, 1450–1457. [Google Scholar] [CrossRef]
- Muñoz-Atienza, E. Systemic and Mucosal B and T Cell Responses Upon Mucosal Vaccination of Teleost Fish. Front. Immunol. 2021, 11, 622377. [Google Scholar] [CrossRef]
- Kumar, S.; Choubey, A.K.; Srivastava, P.K. The Effects of Dietary Immunostimulants on the Innate Immune Response of Indian Major Carp: A Review. Fish Shellfish Immunol. 2022, 123, 36–49. [Google Scholar] [CrossRef]
- Du, Y.; Hu, X.; Miao, L.; Chen, J. Current Status and Development Prospects of Aquatic Vaccines. Front. Immunol. 2022, 13, 1040336. [Google Scholar] [CrossRef]
- Kim, K.-H.; Kim, H.C.; Park, C.-J.; Park, J.-W.; Lee, Y.M.; Kim, W.-J. Interleukin-8 (IL-8) Expression in the Olive Flounder (Paralichthys Olivaceus) against Viral Hemorrhagic Septicemia Virus (VHSV) Challenge. Dev. Reprod. 2019, 23, 231–238. [Google Scholar] [CrossRef]
- Oehlers, S.H.B.; Flores, M.V.; Hall, C.J.; O’Toole, R.; Swift, S.; Crosier, K.E.; Crosier, P.S. Expression of Zebrafish Cxcl8 (Interleukin-8) and Its Receptors during Development and in Response to Immune Stimulation. Dev. Comp. Immunol. 2010, 34, 352–359. [Google Scholar] [CrossRef] [PubMed]
- Buchmann, K.; Karami, A.M.; Duan, Y. The Early Ontogenetic Development of Immune Cells and Organs in Teleosts. Fish Shellfish Immunol. 2024, 146, 109371. [Google Scholar] [CrossRef] [PubMed]
- Grace, M.F.; Manning, M.J. Histogenesis of the Lymphoid Organs in Rainbow Trout, Salmo Gairdneri Rich. 1836. Dev. Comp. Immunol. 1980, 4, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Samanta, A.K.; Oppenheim, J.J.; Matsushima, K. Identification and Characterization of Specific Receptors for Monocyte-Derived Neutrophil Chemotactic Factor (MDNCF) on Human Neutrophils. J. Exp. Med. 1989, 169, 1185–1189. [Google Scholar] [CrossRef]
- Hartl, D.; Latzin, P.; Hordijk, P.; Marcos, V.; Rudolph, C.; Woischnik, M.; Krauss-Etschmann, S.; Koller, B.; Reinhardt, D.; Roscher, A.A.; et al. Cleavage of CXCR1 on Neutrophils Disables Bacterial Killing in Cystic Fibrosis Lung Disease. Nat. Med. 2007, 13, 1423–1430. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santana, P.A.; Forero, J.C.; Guzmán, F.; Gaete, S.; Acosta, F.; Mercado, L.A.; Álvarez, C.A. Detection and Localization of IL-8 and CXCR1 in Rainbow Trout Larvae in Response to Pseudomonas aeruginosa Lipopolysaccharide. Animals 2024, 14, 2878. https://doi.org/10.3390/ani14192878
Santana PA, Forero JC, Guzmán F, Gaete S, Acosta F, Mercado LA, Álvarez CA. Detection and Localization of IL-8 and CXCR1 in Rainbow Trout Larvae in Response to Pseudomonas aeruginosa Lipopolysaccharide. Animals. 2024; 14(19):2878. https://doi.org/10.3390/ani14192878
Chicago/Turabian StyleSantana, Paula A., Juan C. Forero, Fanny Guzmán, Sandra Gaete, Félix Acosta, Luis A. Mercado, and Claudio A. Álvarez. 2024. "Detection and Localization of IL-8 and CXCR1 in Rainbow Trout Larvae in Response to Pseudomonas aeruginosa Lipopolysaccharide" Animals 14, no. 19: 2878. https://doi.org/10.3390/ani14192878
APA StyleSantana, P. A., Forero, J. C., Guzmán, F., Gaete, S., Acosta, F., Mercado, L. A., & Álvarez, C. A. (2024). Detection and Localization of IL-8 and CXCR1 in Rainbow Trout Larvae in Response to Pseudomonas aeruginosa Lipopolysaccharide. Animals, 14(19), 2878. https://doi.org/10.3390/ani14192878






