Metabolomics Reveals the Mechanism by Which Sodium Butyrate Promotes the Liver Pentose Phosphate Pathway and Fatty Acid Synthesis in Lactating Goats
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Experimental Procedures
2.2. Rumen Fluid Collection and Analysis
2.3. Sample Collection
2.4. Liver Metabolite Extraction
2.5. UPLC–MS/MS Analysis
2.6. Data Processing
2.7. RNA Isolation, cDNA Synthesis, and Real-Time PCR
2.8. Western Blot Analysis
2.9. Statistical Analysis
3. Results
3.1. Effect of Sodium Butyrate on the Milk Composition of Lactating Goats Fed a High-Concentrate Diet
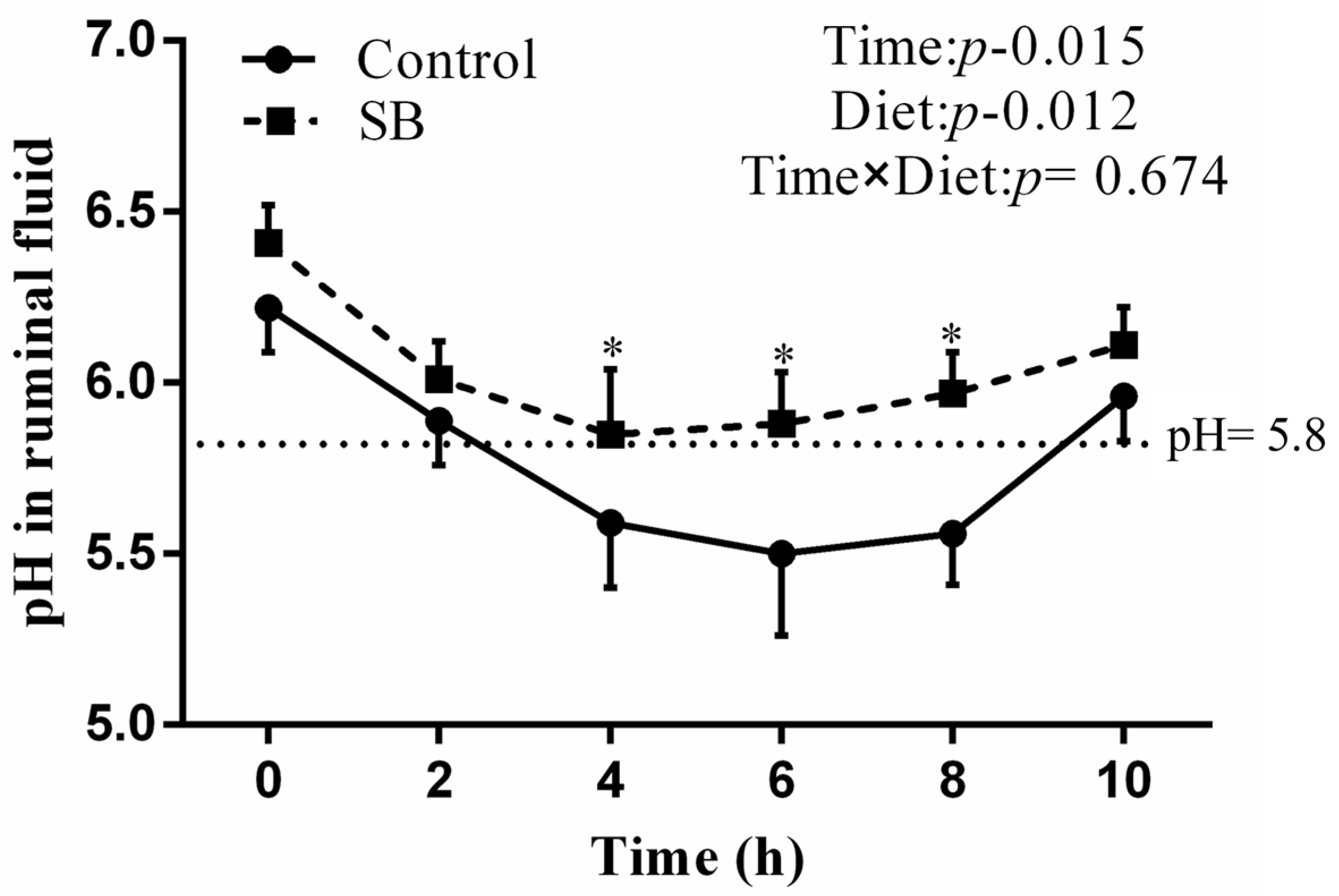
3.2. Rumen pH in the Rumen
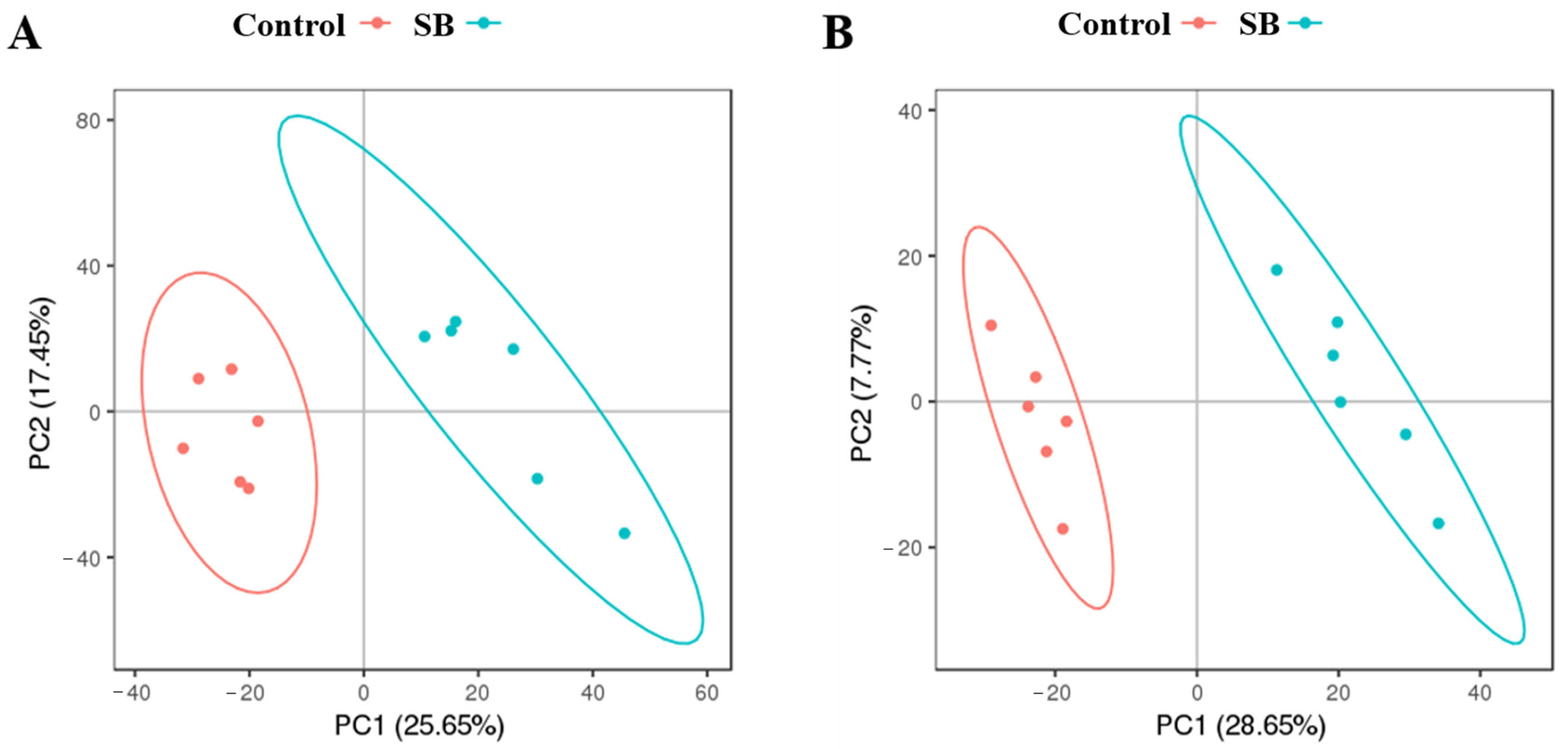
3.3. Analysis of the Liver Metabolism Patterns in Lactating Goats Fed a HC Diet with Sodium Butyrate
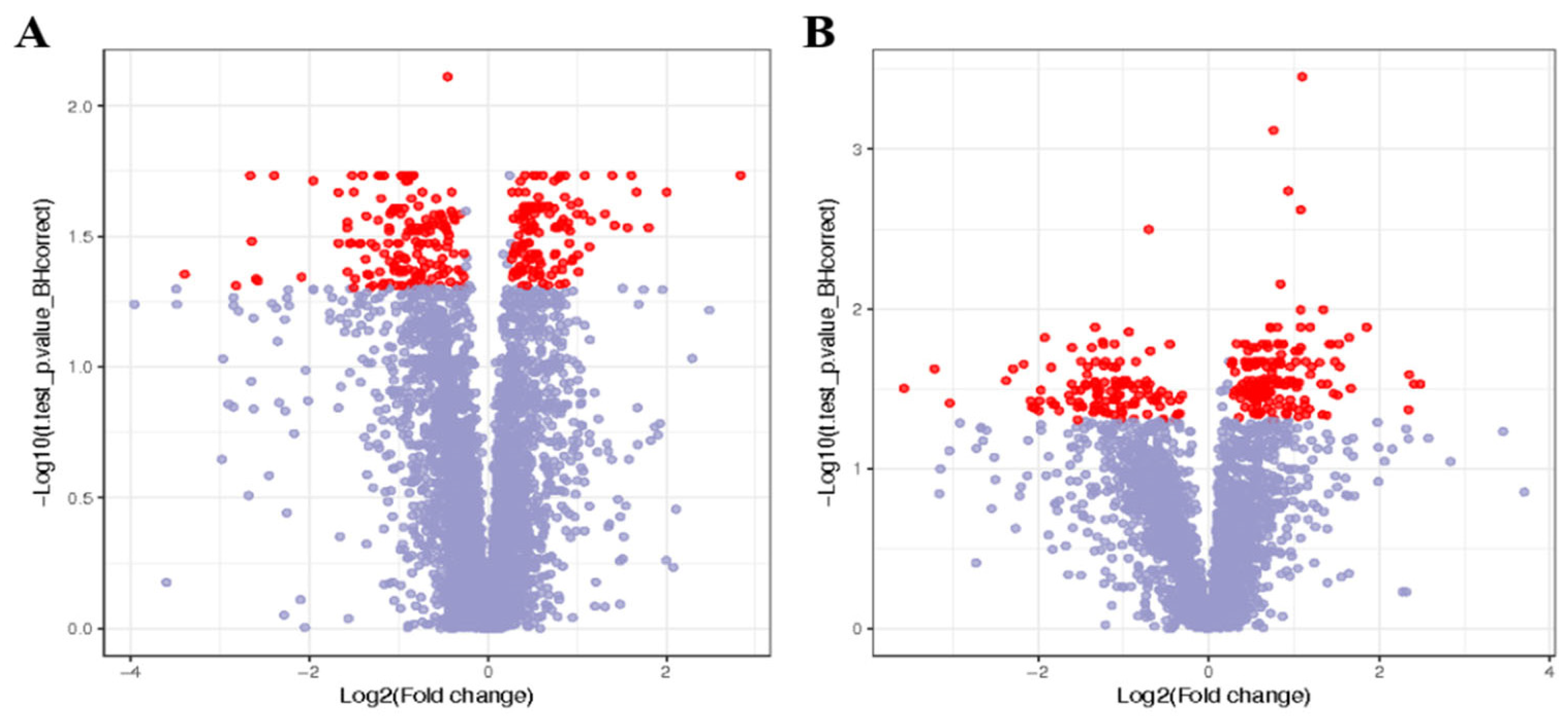
3.4. Screening of Differentially Abundant Metabolites in the Liver
3.5. Pathway Analysis of Differentially Abundant Liver Metabolites
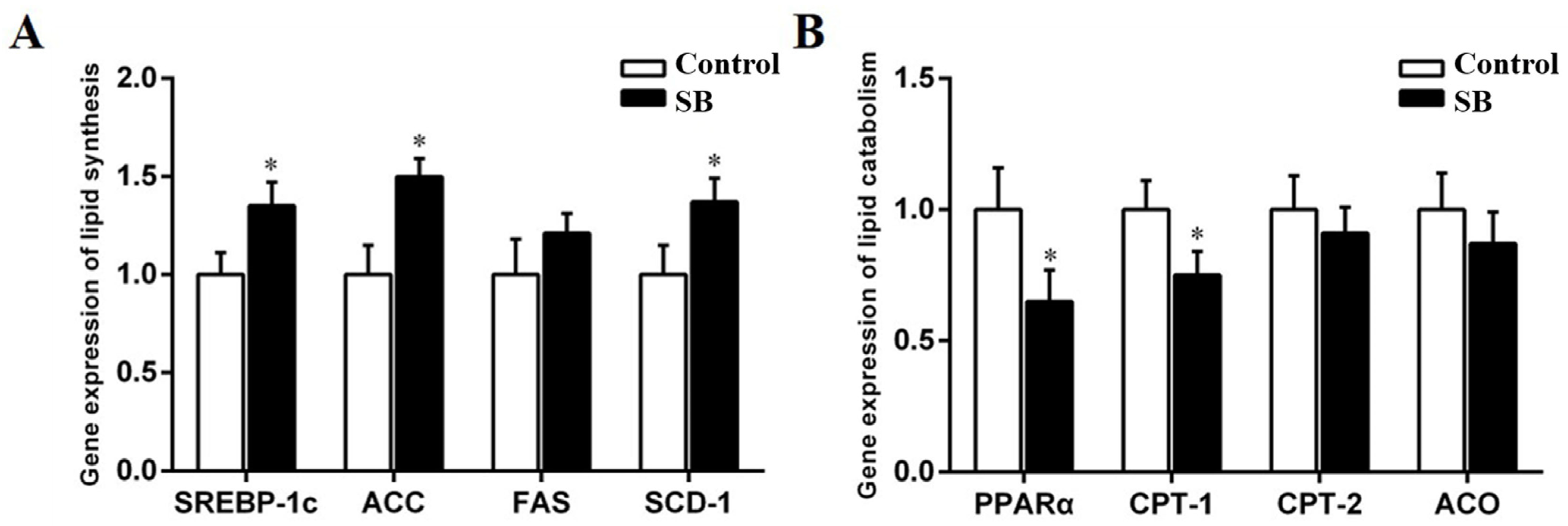
3.6. Sodium Butyrate Treatment Regulated Key Enzymes Involved in Lipid Metabolism in the Livers of Goats
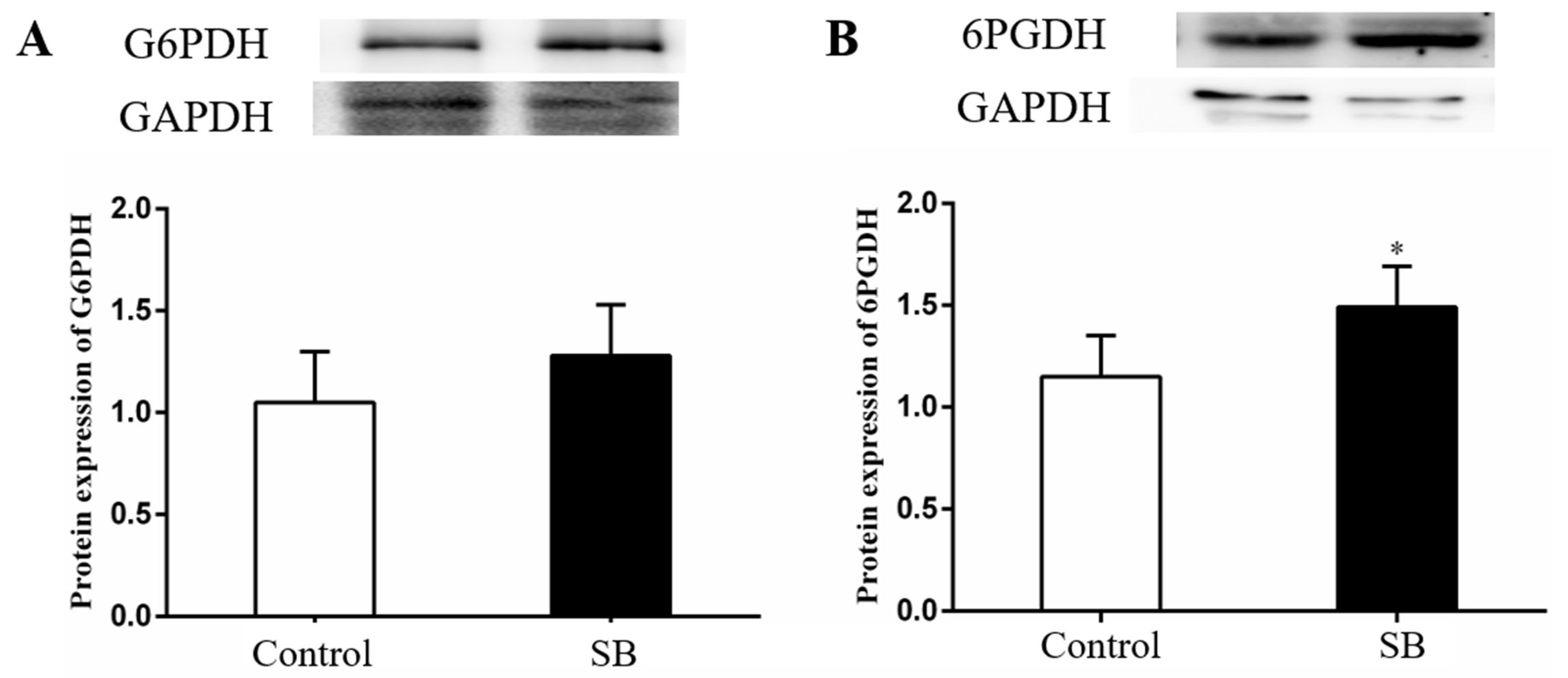
3.7. Sodium Butyrate Treatment Regulated Key Enzymes of the Pentose Phosphate Pathway in the Livers of Goats
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dai, H.; Ma, N.; Chang, G.; Aabdin, Z.U.; Shen, X. Long-term high-concentrate diet feeding induces apoptosis of rumen epithelial cells and inflammation of rumen epithelium in dairy cows. Anim. Biotechnol. 2022, 33, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Huot, F.; Claveau, S.; Bunel, A.; Santschi, D.E.; Gervais, R.; Paquet, É.R. Relationship between farm management strategies, reticuloruminal pH variations, and risks of subacute ruminal acidosis. J. Dairy Sci. 2023, 106, 2487–2497. [Google Scholar] [CrossRef] [PubMed]
- Mikulak, J.; Bruni, E.; Oriolo, F.; Vito, C.D.; Mavilio, D. Hepatic Natural Killer Cells: Organ-Specific Sentinels of Liver Immune Homeostasis and Physiopathology. Front. Immunol. 2019, 10, 946. [Google Scholar] [CrossRef] [PubMed]
- Chang, G.; Zhang, K.; Xu, T.; Jin, D.; Seyfert, H.M.; Shen, X.; Zhuang, S. Feeding a high-grain diet reduces the percentage of LPS clearance and enhances immune gene expression in goat liver. BMC Vet. Res. 2015, 11, 67. [Google Scholar] [CrossRef]
- Dong, H.; Sun, L.; Cong, R.; Tao, S.; DuanMu, Y.; Tian, J.; Ni, Y.; Zhao, R. Changes in milk performance and hepatic metabolism in mid-lactating dairy goats after being fed a high concentrate diet for 10 weeks. Animal 2017, 11, 418–425. [Google Scholar] [CrossRef]
- Li, L.; Cao, Y.; Xie, Z.; Zhang, Y. A high-concentrate diet induced milk fat decline via glucagon-mediated activation of AMP-activated protein kinase in dairy cows. Sci. Rep. 2017, 7, 44217. [Google Scholar] [CrossRef]
- Dong, H.; Wang, S.; Jia, Y.; Ni, Y.; Zhang, Y.; Zhuang, S.; Shen, X.; Zhao, R. Long-term effects of subacute ruminal acidosis (SARA) on milk quality and hepatic gene expression in lactating goats fed a high-concentrate diet. PLoS ONE 2013, 8, e82850. [Google Scholar] [CrossRef]
- Li, X.; Wang, C.; Zhu, J.; Lin, Q.; Yu, M.; Wen, J.; Feng, J.; Hu, C. Sodium butyrate ameliorates oxidative stress-induced intestinal epithelium barrier injury and mitochondrial damage through AMPK-mitophagy pathway. Oxid. Med. Cell. Longev. 2022, 2022, 3745135. [Google Scholar] [CrossRef]
- Chen, G.; Ran, X.; Li, B.; Li, Y.; He, D.; Huang, B.; Fu, S.; Liu, J.; Wang, W. Sodium butyrate inhibits inflammation and maintains epithelium barrier integrity in a TNBS-induced inflammatory bowel disease mice model. EBioMedicine 2018, 30, 317–325. [Google Scholar] [CrossRef]
- Li, C.; Chen, J.; Zhao, M.; Liu, M.; Yue, Z.; Liu, L.; Li, F. Effect of sodium butyrate on slaughter performance, serum indexes and intestinal barrier of rabbits. J. Anim. Physiol. Anim. Nutr. 2022, 106, 156–166. [Google Scholar] [CrossRef]
- Sun, Q.; Ji, Y.C.; Wang, Z.L.; She, X.; He, Y.; Ai, Q.; Li, L.Q. Sodium butyrate alleviates intestinal inflammation in mice with necrotizing enterocolitis. Mediat. Inflamm. 2021, 2021, 6259381. [Google Scholar] [CrossRef]
- Zhang, J.; Gaowa, N.; Wang, Y.; Li, H.; Cao, Z.; Yang, H.; Zhang, X.; Li, S. Complementary hepatic metabolomics and proteomics reveal the adaptive mechanisms of dairy cows to the transition period. J. Dairy Sci. 2023, 106, 2071–2088. [Google Scholar] [CrossRef]
- Ghaffari, M.; Daniel, J.; Sadri, H.; Schuchardt, S.; Martín-Tereso, J.; Sauerwein, H. Longitudinal characterization of the metabolome of dairy cows transitioning from one lactation to the next: Investigations in blood serum. J. Dairy Sci. 2024, 107, 1263–1285. [Google Scholar] [CrossRef]
- Chen, M.; Xie, W.; Zhou, S.; Ma, N.; Wang, Y.; Huang, J.; Shen, X.; Chang, G. A high-concentrate diet induces colonic inflammation and barrier damage in Hu sheep. J. Dairy Sci. 2023, 106, 9644–9662. [Google Scholar] [CrossRef]
- Plaizier, J.C.; Mulligan, F.J.; Neville, E.W.; Guan, L.L.; Steele, M.A.; Penner, G.B. Invited review: Effect of subacute ruminal acidosis on gut health of dairy cows. J. Dairy Sci. 2022, 105, 7141–7160. [Google Scholar] [CrossRef]
- Artegoitia, V.M.; Foote, A.P.; Lewis, R.M.; Freetly, H.C. Rumen fluid metabolomics analysis associated with feed efficiency on crossbred steers. Sci. Rep. 2017, 7, 2864. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, Z.; Ye, B.; Ma, J.H.; Ji, S.; Sheng, W.; Ye, S.; Ou, Y.; Peng, Y.; Yang, X. Sodium butyrate ameliorates diabetic retinopathy in mice via the regulation of gut microbiota and related short-chain fatty acids. J. Transl. Med. 2023, 21, 451. [Google Scholar] [CrossRef]
- Cheng, J.; Zhang, Y.; Ge, Y.; Li, W.; Cao, Y.; Qu, Y.; Liu, S.; Guo, Y.; Fu, S.; Liu, J. Sodium butyrate promotes milk fat synthesis in bovine mammary epithelial cells via GPR41 and its downstream signalling pathways. Life Sci. 2020, 259, 118375. [Google Scholar] [CrossRef]
- Zhao, H.; Bai, H.; Deng, F.; Zhong, R.; Liu, L.; Chen, L.; Zhang, H. Chemically protected sodium butyrate improves growth performance and early development and function of small intestine in broilers as one effective substitute for antibiotics. Antibiotics 2022, 11, 132. [Google Scholar] [CrossRef]
- Ali, I.; Li, C.; Kuang, M.; Shah, A.U.; Shafiq, M.; Ahmad, M.A.; Abdalmegeed, D.; Li, L.; Wang, G. Nrf2 Activation and NF-Kb & caspase/bax signaling inhibition by sodium butyrate alleviates LPS-induced cell injury in bovine mammary epithelial cells. Mol. Immunol. 2022, 148, 54–67. [Google Scholar]
- Meng, M.; Li, X.; Huo, R.; Chang, G.; Shen, X. Effects of dietary disodium fumarate supplementation on muscle quality, chemical composition, oxidative stress and lipid metabolism of Hu sheep induced by high concentrate diet. Meat Sci. 2023, 201, 109176. [Google Scholar] [CrossRef]
- Zhang, J.; Bu, L.; Liu, Y.; Huo, W.; Xia, C.; Pei, C.; Liu, Q. Dietary supplementation of sodium butyrate enhances lactation performance by promoting nutrient digestion and mammary gland development in dairy cows. Anim. Nutr. 2023, 15, 137–148. [Google Scholar] [CrossRef]
- Fukumori, R.; Doi, K.; Mochizuki, T.; Oikawa, S.; Gondaira, S.; Iwasaki, T.; Izumi, K. Sodium butyrate administration modulates the ruminal villus height, inflammation-related gene expression, and plasma hormones concentration in dry cows fed a high-fiber diet. Anim. Sci. J. 2022, 93, e13791. [Google Scholar] [CrossRef]
- Ma, N.; Abaker, J.A.; Wei, G.; Chen, H.; Shen, X.; Chang, G. A high-concentrate diet induces an inflammatory response and oxidative stress and depresses milk fat synthesis in the mammary gland of dairy cows. J. Dairy Sci. 2022, 105, 5493–5505. [Google Scholar] [CrossRef]
- Bauman, D.E.; Harvatine, K.J.; Lock, A.L. Nutrigenomics, rumen-derived bioactive fatty acids, and the regulation of milk fat synthesis. Annu. Rev. Nutr. 2011, 31, 299–319. [Google Scholar] [CrossRef]
- Moyes, K.M.; Sørensen, P.; Bionaz, M. The Impact of Intramammary Escherichia coli Challenge on Liver and Mammary Transcriptome and Cross-Talk in Dairy Cows during Early Lactation Using RNAseq. PLoS ONE 2016, 11, e0157480. [Google Scholar] [CrossRef]
- Kalinowski, C.T.; Betancor, M.B.; Torrecillas, S.; Sprague, M.; Larroquet, L.; Véron, V.; Panserat, S.; Izquierdo, M.S.; Kaushik, S.J.; Fontagné-Dicharry, S. More than an antioxidant: Role of dietary astaxanthin on lipid and glucose metabolism in the liver of rainbow trout (Oncorhynchus mykiss). Antioxidants 2023, 12, 136. [Google Scholar] [CrossRef]
- Li, L.; Zhang, X.; Ren, H.; Huang, X.; Shen, T.; Tang, W.; Dou, L.; Li, J. miR-23a/b-3p promotes hepatic lipid accumulation by regulating Srebp-1c and Fas. J. Mol. Endocrinol. 2022, 68, 35–49. [Google Scholar] [CrossRef]
- Mun, J.; Kim, S.; Yoon, H.G.; You, Y.; Kim, O.K.; Choi, K.C.; Lee, Y.H.; Lee, J.; Park, J.; Jun, W. Water extract of Curcuma longa L. ameliorates non-alcoholic fatty liver disease. Nutrients 2019, 11, 2536. [Google Scholar] [CrossRef]
- Kong, L.; Zhang, W.; Liu, S.; Zhong, Z.; Zheng, G. Quercetin, Engelitin and Caffeic Acid of Smilax china L. Polyphenols, Stimulate 3T3-L1 Adipocytes to Brown-like Adipocytes via β 3-AR/AMPK Signaling Pathway. Plant Foods Hum. Nutr. 2022, 77, 529–537. [Google Scholar] [CrossRef] [PubMed]
- Honma, K.; Oshima, K.; Takami, S.; Goda, T. Regulation of hepatic genes related to lipid metabolism and antioxidant enzymes by sodium butyrate supplementation. Metab. Open 2020, 7, 100043. [Google Scholar] [CrossRef]
- Tan, V.W.; Salmi, T.M.; Karamalakis, A.P.; Gillespie, A.; Ong, A.J.S.; Balic, J.J.; Chan, Y.C.; Bladen, C.E.; Brown, K.K.; Dawson, M.A. SLAM-ITseq identifies that Nrf2 induces liver regeneration through the pentose phosphate pathway. Dev. Cell. 2024, 59, 898–910. [Google Scholar] [CrossRef]
- Fuentes-Lemus, E.; Reyes, J.S.; Figueroa, J.D.; Davies, M.J.; López-Alarcón, C. The enzymes of the oxidative phase of the pentose phosphate pathway as targets of reactive species: Consequences for NADPH production. Biochem. Soc. Trans. 2023, 51, 2173–2187. [Google Scholar] [CrossRef]
- Dobrowolski, A.; Mirończuk, A.M. The influence of transketolase on lipid biosynthesis in the yeast Yarrowia lipolytica. Microb. Cell Fact. 2020, 19, 138. [Google Scholar] [CrossRef]
- Wang, D.; Guo, M.; Li, X.; Zhao, D.; Wang, M. Microbiota, co-metabolites, and network pharmacology reveal the alteration of the ginsenoside fraction on inflammatory bowel disease. J. Ginseng Res. 2023, 47, 54–64. [Google Scholar] [CrossRef]

| Concentrate: Forage Ratio 60:40 | |||
|---|---|---|---|
| Ingredient (% of Dry Matter) | Nutrient Level b | ||
| Leymus chinensis | 27.00 | Net energy/(MJ·kg−1) | 6.71 |
| Alfalfa silage | 13.00 | Crude protein/% | 16.92 |
| Corn | 23.24 | Neutral detergent fibre/% | 31.45 |
| Wheat bran | 20.77 | Acid detergent fibre/% | 17.56 |
| Soybean meal | 13.67 | Calcium/% | 0.89 |
| Limestone | 1.42 | Phosphorus/% | 0.46 |
| NaCl | 0.30 | ||
| Premix a | 0.60 | ||
| Total | 100.00 | ||
| Target Genes | Primer Sequences (5′-3′) | Products/bp |
|---|---|---|
| GAPDH | GGGTCATCATCTCTGCACCT GGTCATAAGTCCCTCCACGA | 177 |
| ACC | ACGCAGGCATCAGAAGATTA GAGGGTTCAGTTCCAGAAAGTA | 179 |
| FAS | GCACTACCACAACCCAAACCC CGTTGGAGCCACCGAAGC | 161 |
| SCD-1 | CCGCCCTGAAATGAGAGATG AGGGCTCCCAAGTGTAACAGAC | 154 |
| SREBP-1c | CGACTACATCCGCTTCCTTCA ACTTCCACCGCTGCTACTG | 259 |
| CPT-1 | CCCATGTCCTTGTAATGAGCCAG AGACTTCGCTGAGCAGTGCCA | 230 |
| CPT-2 | ACGCCGTGAAGTATAACCCT CCAAAAATCGCTTGTCCCTT | 119 |
| ACO | TAAGCCTTTGCCAGGTATT ATGGTCCCGTAGGTCAG | 189 |
| PPARα | GGAGGTCCGCATCTTCCACT GCAGCAAATGATAGCAGCCACA | 352 |
| Control | SB | p Value | |
|---|---|---|---|
| Milk yield, g/d | 988.25 ± 31.34 | 1024.14 ± 26.18 | 0.136 |
| Milk protein | |||
| % | 2.82 ± 0.11 | 3.04 ± 0.02 | 0.086 |
| g/d | 29.35 ± 1.24 | 30.04 ± 0.35 | 0.479 |
| Milk fat | |||
| % | 3.02 ± 0.07 | 3.69 ± 0.14 * | 0.015 |
| g/d | 34.46 ± 0.23 | 37.35 ± 0.73 * | 0.048 |
| Mode | Metabolites | FC | p Value | VIP | Trend | Metabolic Pathways |
|---|---|---|---|---|---|---|
| POS | Bovinic acid | 1.45 | 0.01 | 1.61 | up | Linoleic acid metabolism |
| Linoleic acid | 1.45 | 0.01 | 1.61 | up | Linoleic acid metabolism | |
| Palmitoleic acid | 3.04 | 0.01 | 2.77 | up | Fatty acid biosynthesis | |
| Dodecanoic acid | 0.59 | 0.03 | 1.81 | down | Fatty acid biosynthesis | |
| Eicosadienoic acid | 1.60 | 0.02 | 1.66 | up | Biosynthesis of unsaturated fatty acids | |
| Docosapentaenoic acid | 1.37 | 0.03 | 1.46 | up | Biosynthesis of unsaturated fatty acids | |
| Glycerophosphocholine | 1.25 | 0.04 | 1.14 | up | Glycerophospholipid metabolism | |
| Phosphorylcholine | 1.36 | 0.04 | 1.26 | up | Glycerophospholipid metabolism | |
| LysoPC (18:3 (6Z, 9Z, 12Z)) | 0.71 | 0.02 | 1.35 | up | Glycerophospholipid metabolism | |
| LysoPC (22:4 (7Z, 10Z, 13Z, 16Z)) | 0.52 | 0.04 | 2.08 | down | Glycerophospholipid metabolism | |
| LysoPC (16:0) | 0.71 | 0.02 | 1.35 | down | Glycerophospholipid metabolism | |
| LysoPC (18:3 (9Z, 12Z, 15Z)) | 0.71 | 0.02 | 1.35 | down | Glycerophospholipid metabolism | |
| Xylulose 5-phosphate | 1.36 | 0.02 | 1.42 | up | Pentose phosphate pathway | |
| d-Ribulose 5-phosphate | 1.36 | 0.02 | 1.42 | up | Pentose phosphate pathway | |
| Ribose 1-phosphate | 1.36 | 0.02 | 1.42 | up | Pentose phosphate pathway | |
| d-Ribose 5-phosphate | 1.36 | 0.02 | 1.42 | up | Pentose phosphate pathway | |
| NEG | LysoPC (20:4 (8Z, 11Z, 14Z, 17Z)) | 0.59 | 0.03 | 1.45 | down | Glycerophospholipid metabolism |
| LysoPC (20:4 (5Z, 8Z, 11Z, 14Z)) | 0.47 | 0.034 | 1.78 | down | Glycerophospholipid metabolism | |
| PE (14:1 (9Z)/14:1 (9Z)) | 0.10 | 0.02 | 2.92 | down | Glycerophospholipid metabolism | |
| Galactosylglycerol | 2.28 | 0.01 | 1.85 | up | Glycerolipid metabolism | |
| Eicosapentaenoic acid | 1.49 | 0.03 | 1.28 | up | Biosynthesis of unsaturated fatty acids | |
| 6-Phosphogluconic acid | 1.78 | 0.02 | 1.38 | up | Pentose phosphate pathway | |
| Adenosine monophosphate | 1.59 | 0.03 | 1.30 | up | Regulation of lipolysis in adipocytes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; Chen, X.; Yan, S.; Zhang, Y. Metabolomics Reveals the Mechanism by Which Sodium Butyrate Promotes the Liver Pentose Phosphate Pathway and Fatty Acid Synthesis in Lactating Goats. Animals 2024, 14, 3249. https://doi.org/10.3390/ani14223249
Li L, Chen X, Yan S, Zhang Y. Metabolomics Reveals the Mechanism by Which Sodium Butyrate Promotes the Liver Pentose Phosphate Pathway and Fatty Acid Synthesis in Lactating Goats. Animals. 2024; 14(22):3249. https://doi.org/10.3390/ani14223249
Chicago/Turabian StyleLi, Lin, Xi Chen, Shuping Yan, and Yuanshu Zhang. 2024. "Metabolomics Reveals the Mechanism by Which Sodium Butyrate Promotes the Liver Pentose Phosphate Pathway and Fatty Acid Synthesis in Lactating Goats" Animals 14, no. 22: 3249. https://doi.org/10.3390/ani14223249
APA StyleLi, L., Chen, X., Yan, S., & Zhang, Y. (2024). Metabolomics Reveals the Mechanism by Which Sodium Butyrate Promotes the Liver Pentose Phosphate Pathway and Fatty Acid Synthesis in Lactating Goats. Animals, 14(22), 3249. https://doi.org/10.3390/ani14223249




