Spotting the Pattern: A Review on White Coat Color in the Domestic Horse
Abstract
:Simple Summary
Abstract
1. Introduction
2. Dominant White
2.1. Phenotype
2.2. Mechanisms and Genetics
3. Tobiano, Sabino, and Roan
3.1. Tobiano
3.1.1. Phenotype
3.1.2. Genetics and Mechanism
3.2. Sabino
3.2.1. Phenotype
3.2.2. Genetics and Mechanism
3.3. Roan
4. Splashed White
4.1. Phenotype
4.2. MITF
4.3. PAX3
5. Eden White and HPS5
6. Lethal White Overo
7. Leopard Complex Spotting
7.1. Congenital Stationary Night Blindness
7.2. Equine Recurrent Uveitis
7.3. RFWD3 and PATN1
8. Conclusions and Future Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bailey, E.; Brooks, S.A. Horse Genetics, 3rd ed.; CABI: Wallingford, UK, 2020. [Google Scholar]
- Haase, B.; Brooks, S.A.; Schlumbaum, A.; Azor, P.J.; Bailey, E.; Alaeddine, F.; Mevissen, M.; Burger, D.; Poncet, P.-A.; Rieder, S.; et al. Allelic Heterogeneity at the Equine KIT Locus in Dominant White (W) Horses. PLoS Genet. 2007, 3, e195. [Google Scholar] [CrossRef]
- Grilz-Seger, G.; Neuditschko, M.; Ricard, A.; Velie, B.; Lindgren, G.; Mesarič, M.; Cotman, M.; Horna, M.; Dobretsberger, M.; Brem, G.; et al. Genome-Wide Homozygosity Patterns and Evidence for Selection in a Set of European and Near Eastern Horse Breeds. Genes 2019, 10, 491. [Google Scholar] [CrossRef]
- He, Z.; Dai, X.; Lyu, W.; Beaumont, M.; Yu, F. Estimating Temporally Variable Selection Intensity from Ancient DNA Data. Mol. Biol. Evol. 2023, 40, msad008. [Google Scholar] [CrossRef]
- Esdaile, E.; Till, B.; Kallenberg, A.; Fremeux, M.; Bickel, L.; Bellone, R.R. A de novo missense mutation in KIT is responsible for dominant white spotting phenotype in a Standardbred horse. Anim. Genet. 2022, 53, 534–537. [Google Scholar] [CrossRef]
- McFadden, A.; Martin, K.; Foster, G.; Vierra, M.; Lundquist, E.W.; Everts, R.E.; Martin, E.; Volz, E.; McLoone, K.; Brooks, S.A.; et al. 5′UTR Variant in KIT Associated With White Spotting in Horses. J. Equine Vet. Sci. 2023, 127, 104563. [Google Scholar] [CrossRef]
- Patterson Rosa, L.; Martin, K.; Vierra, M.; Foster, G.; Lundquist, E.; Brooks, S.A.; Lafayette, C. Two Variants of KIT Causing White Patterning in Stock-Type Horses. J. Hered. 2021, 112, 447–451. [Google Scholar] [CrossRef] [PubMed]
- Patterson Rosa, L.; Martin, K.; Vierra, M.; Lundquist, E.; Foster, G.; Brooks, S.A.; Lafayette, C. A KIT Variant Associated with Increased White Spotting Epistatic to MC1R Genotype in Horses (Equus caballus). Animals 2022, 12, 1958. [Google Scholar] [CrossRef] [PubMed]
- Brooks, S.A.; Bailey, E. Exon skipping in the KIT gene causes a Sabino spotting pattern in horses. Mamm. Genome 2005, 16, 893–902. [Google Scholar] [CrossRef] [PubMed]
- Capomaccio, S.; Milanesi, M.; Nocelli, C.; Giontella, A.; Verini-Supplizi, A.; Branca, M.; Silvestrelli, M.; Cappelli, K. Splicing site disruption in the KIT gene as strong candidate for white dominant phenotype in an Italian Trotter. Anim. Genet. 2017, 48, 727–728. [Google Scholar] [CrossRef] [PubMed]
- Dürig, N.; Jude, R.; Holl, H.; Brooks, S.A.; Lafayette, C.; Jagannathan, V.; Leeb, T. Whole genome sequencing reveals a novel deletion variant in the KIT gene in horses with white spotted coat colour phenotypes. Anim. Genet. 2017, 48, 483–485. [Google Scholar] [CrossRef] [PubMed]
- Haase, B.; Brooks, S.A.; Tozaki, T.; Burger, D.; Poncet, P.-A.; Rieder, S.; Hasegawa, T.; Penedo, C.; Leeb, T. Seven novel KIT mutations in horses with white coat colour phenotypes. Anim. Genet. 2009, 40, 623–629. [Google Scholar] [CrossRef]
- Haase, B.; Jagannathan, V.; Rieder, S.; Leeb, T. A novel KIT variant in an Icelandic horse with white-spotted coat colour. Anim. Genet. 2015, 46, 466. [Google Scholar] [CrossRef]
- Haase, B.; Rieder, S.; Tozaki, T.; Hasegawa, T.; Penedo, M.C.T.; Jude, R.; Leeb, T. Five novel KIT mutations in horses with white coat colour phenotypes: BRIEF NOTE. Anim. Genet. 2011, 42, 337–339. [Google Scholar] [CrossRef]
- Hauswirth, R.; Jude, R.; Haase, B.; Bellone, R.R.; Archer, S.; Holl, H.; Brooks, S.A.; Tozaki, T.; Penedo, M.C.T.; Rieder, S.; et al. Novel variants in the KIT and PAX3 genes in horses with white-spotted coat colour phenotypes. Anim. Genet. 2013, 44, 763–765. [Google Scholar] [CrossRef]
- Hoban, R.; Castle, K.; Hamilton, N.; Haase, B. Novel KIT variants for dominant white in the Australian horse population. Anim. Genet. 2018, 49, 99–100. [Google Scholar] [CrossRef] [PubMed]
- Holl, H.; Brooks, S.; Bailey, E. De novo mutation of KIT discovered as a result of a non-hereditary white coat colour pattern: De novo mutation of KIT. Anim. Genet. 2010, 41, 196–198. [Google Scholar] [CrossRef]
- Holl, H.M.; Brooks, S.A.; Carpenter, M.L.; Bustamante, C.D.; Lafayette, C. A novel splice mutation within equine KIT and the W15 allele in the homozygous state lead to all white coat color phenotypes. Anim. Genet. 2017, 48, 497–498. [Google Scholar] [CrossRef]
- Hug, P.; Jude, R.; Henkel, J.; Jagannathan, V.; Leeb, T. A novel KIT deletion variant in a German Riding Pony with white-spotting coat colour phenotype. Anim. Genet. 2019, 50, 761–763. [Google Scholar] [CrossRef]
- Martin, K.; Patterson Rosa, L.; Vierra, M.; Foster, G.; Brooks, S.A.; Lafayette, C. De novo mutation of KIT causes extensive coat white patterning in a family of Berber horses. Anim. Genet. 2021, 52, 135–137. [Google Scholar] [CrossRef] [PubMed]
- Esdaile, E.; Kallenberg, A.; Avila, F.; Bellone, R.R. Identification of W13 in the American Miniature Horse and Shetland Pony Populations. Genes 2021, 12, 1985. [Google Scholar] [CrossRef] [PubMed]
- Nicholas, F.; Tammen, I.; Hub, S.I. Online Mendelian Inheritance in Animals (OMIA). 1995. Available online: https://ses.library.usyd.edu.au/handle/2123/31190 (accessed on 17 July 2023).
- Avila, F.; Hughes, S.S.; Magdesian, K.G.; Penedo, M.C.T.; Bellone, R.R. Breed Distribution and Allele Frequencies of Base Coat Color, Dilution, and White Patterning Variants across 28 Horse Breeds. Genes 2022, 13, 1641. [Google Scholar] [CrossRef]
- Pasternak, M.; Krupiński, J.; Gurgul, A.; Bugno-Poniewierska, M. Genetic, historical and breeding aspects of the occurrence of the tobiano pattern and white markings in the Polish population of Hucul horses–A review. J. Appl. Anim. Res. 2020, 48, 21–27. [Google Scholar] [CrossRef]
- Pulos, W.L.; Hutt, F.B. Lethal Dominant White in Horses. J. Hered. 1969, 60, 59–63. [Google Scholar] [CrossRef] [PubMed]
- McFadden, A.; Vierra, M.; Robilliard, H.; Martin, K.; Brooks, S.A.; Everts, R.E.; Lafayette, C. Population Analysis Identifies 15 Multi-Variant Dominant White Haplotypes in Horses. Animals, 2023; Manuscript submitted for review. [Google Scholar]
- Bellone, R.R.; Holl, H.; Setaluri, V.; Devi, S.; Maddodi, N.; Archer, S.; Sandmeyer, L.; Ludwig, A.; Foerster, D.; Pruvost, M.; et al. Evidence for a retroviral insertion in TRPM1 as the cause of congenital stationary night blindness and leopard complex spotting in the horse. PLoS ONE 2013, 8, e78280. [Google Scholar] [CrossRef] [PubMed]
- Phung, B.; Sun, J.; Schepsky, A.; Steingrimsson, E.; Rönnstrand, L. C-KIT Signaling Depends on Microphthalmia-Associated Transcription Factor for Effects on Cell Proliferation. PLoS ONE 2011, 6, e24064. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.J.; Erickson, C.A. The making of a melanocyte: The specification of melanoblasts from the neural crest. Pigment. Cell Melanoma Res. 2008, 21, 598–610. [Google Scholar] [CrossRef] [PubMed]
- Fuse, N.; Yasumoto, K.; Suzuki, H.; Takahashi, K.; Shibahara, S. Identification of a Melanocyte-Type Promoter of the Microphthalmia-Associated Transcription Factor Gene. Biochem. Biophys. Res. Commun. 1996, 219, 702–707. [Google Scholar] [CrossRef]
- Jones, S. An overview of the basic helix-loop-helix proteins. Genome Biol. 2004, 5, 226. [Google Scholar] [CrossRef]
- Kawakami, A.; Fisher, D.E. The master role of microphthalmia-associated transcription factor in melanocyte and melanoma biology. Lab. Investig. 2017, 97, 649–656. [Google Scholar] [CrossRef]
- Williams, D.E.; de Vries, P.; Namen, A.E.; Widmer, M.B.; Lyman, S.D. The Steel factor. Dev. Biol. 1992, 151, 368–376. [Google Scholar] [CrossRef]
- Yasumoto, K.; Yokoyama, K.; Shibata, K.; Tomita, Y.; Shibahara, S. Microphthalmia-associated transcription factor as a regulator for melanocyte-specific transcription of the human tyrosinase gene. Mol. Cell. Biol. 1994, 14, 8058–8070. [Google Scholar] [CrossRef]
- Yutzey, K.E.; Konieczny, S.F. Different E-box regulatory sequences are functionally distinct when placed within the context of the troponin I enhancer. Nucleic Acids Res. 1992, 20, 5105–5113. [Google Scholar] [CrossRef] [PubMed]
- Brooks, S.A.; Lear, T.L.; Adelson, D.L.; Bailey, E. A chromosome inversion near the KIT gene and the Tobiano spotting pattern in horses. Cytogenet. Genome Res. 2007, 119, 225–230. [Google Scholar] [CrossRef] [PubMed]
- Grilz-Seger, G.; Reiter, S.; Neuditschko, M.; Wallner, B.; Rieder, S.; Leeb, T.; Jagannathan, V.; Mesarič, M.; Cotman, M.; Pausch, H.; et al. A Genome-Wide Association Analysis in Noriker Horses Identifies a SNP Associated With Roan Coat Color. J. Equine Vet. Sci. 2020, 88, 102950. [Google Scholar] [CrossRef] [PubMed]
- Hintz, H.F.; van Vleck, L.D. Lethal dominant roan in horses. J. Hered. 1979, 70, 145–146. [Google Scholar] [CrossRef]
- Marklund, S.; Moller, M.; Sandberg, K.; Andersson, L. Close association between sequence polymorphism in the KIT gene and the roan coat color in horses. Mamm. Genome 1999, 10, 283–288. [Google Scholar] [CrossRef]
- Ludwig, A.; Pruvost, M.; Reissmann, M.; Benecke, N.; Brockmann, G.A.; Castaños, P.; Cieslak, M.; Lippold, S.; Llorente, L.; Malaspinas, A.-S.; et al. Coat color variation at the beginning of horse domestication. Science 2009, 324, 485. [Google Scholar] [CrossRef]
- Stachurska, A.; Jansen, P. Crypto-tobiano horses in Hucul breed. Czech J. Anim. Sci. 2015, 60, 1–9. [Google Scholar] [CrossRef]
- Kawakami, T.; Jensen, M.K.; Slavney, A.; Deane, P.E.; Milano, A.; Raghavan, V.; Ford, B.; Chu, E.T.; Sams, A.J.; Boyko, A.R. R-locus for roaned coat is associated with a tandem duplication in an intronic region of USH2A in dogs and also contributes to Dalmatian spotting. PLoS ONE 2021, 16, e0248233. [Google Scholar] [CrossRef]
- Bellone, R.R.; Tanaka, J.; Esdaile, E.; Sutton, R.B.; Payette, F.; Leduc, L.; Till, B.J.; Abdel-Ghaffar, A.K.; Hammond, M.; Magdesian, K.G. A de novo 2.3 kb structural variant in MITF explains a novel splashed white phenotype in a Thoroughbred family. Anim. Genet. 2023, 54, 752–762. [Google Scholar] [CrossRef]
- Hauswirth, R.; Haase, B.; Blatter, M.; Brooks, S.A.; Burger, D.; Drögemüller, C.; Gerber, V.; Henke, D.; Janda, J.; Jude, R.; et al. Mutations in MITF and PAX3 Cause “Splashed White” and Other White Spotting Phenotypes in Horses. PLoS Genet. 2012, 8, e1002653. [Google Scholar] [CrossRef]
- Henkel, J.; Lafayette, C.; Brooks, S.A.; Martin, K.; Patterson-Rosa, L.; Cook, D.; Jagannathan, V.; Leeb, T. Whole-genome sequencing reveals a large deletion in the MITF gene in horses with white spotted coat colour and increased risk of deafness. Anim. Genet. 2019, 50, 172–174. [Google Scholar] [CrossRef] [PubMed]
- Magdesian, K.G.; Tanaka, J.; Bellone, R.R. A De Novo MITF Deletion Explains a Novel Splashed White Phenotype in an American Paint Horse. J. Hered. 2020, 111, 287–293. [Google Scholar] [CrossRef]
- McFadden, A.; Martin, K.; Foster, G.; Vierra, M.; Lundquist, E.W.; Everts, R.E.; Martin, E.; Volz, E.; McLoone, K.; Brooks, S.A.; et al. Two Novel Variants in MITF and PAX3 Associated with Splashed White Phenotypes in Horses. J. Equine Vet. Sci. 2023, 128, 104875. [Google Scholar] [CrossRef]
- Patterson Rosa, L.; Martin, K.; Vierra, M.; Foster, G.; Brooks, S.A.; Lafayette, C. Non-frameshift deletion on MITF is associated with a novel splashed white spotting pattern in horses (Equus caballus). Anim. Genet. 2022, 53, 538–540. [Google Scholar] [CrossRef] [PubMed]
- Galibert, M.-D.; Dexter, T.J.; Goding, C.R.; Yavuzer, U. Pax3 and Regulation of the Melanocyte-specific Tyrosinase-related Protein-1 Promoter. J. Biol. Chem. 1999, 274, 26894–26900. [Google Scholar] [CrossRef]
- Pogenberg, V.; Ögmundsdóttir, M.H.; Bergsteinsdóttir, K.; Schepsky, A.; Phung, B.; Deineko, V.; Milewski, M.; Steingrímsson, E.; Wilmanns, M. Restricted leucine zipper dimerization and specificity of DNA recognition of the melanocyte master regulator MITF. Genes Dev. 2012, 26, 2647–2658. [Google Scholar] [CrossRef] [PubMed]
- Dürig, N.; Jude, R.; Jagannathan, V.; Leeb, T. A novel MITF variant in a white American Standardbred foal. Anim. Genet. 2017, 48, 123–124. [Google Scholar] [CrossRef]
- Negro, S.; Imsland, F.; Valera, M.; Molina, A.; Solé, M.; Andersson, L. Association analysis of KIT, MITF, and PAX3 variants with white markings in Spanish horses. Anim. Genet. 2017, 48, 349–352. [Google Scholar] [CrossRef]
- Scott, M.P.; Tamkun, J.W.; Hartzell, G.W. The structure and function of the homeodomain. Biochim. Biophys. Acta (BBA)-Rev. Cancer 1989, 989, 25–48. [Google Scholar] [CrossRef]
- Thompson, B.; Davidson, E.A.; Liu, W.; Nebert, D.W.; Bruford, E.A.; Zhao, H.; Dermitzakis, E.T.; Thompson, D.C.; Vasiliou, V. Overview of PAX gene family: Analysis of human tissue-specific variant expression and involvement in human disease. Hum. Genet. 2021, 140, 381–400. [Google Scholar] [CrossRef]
- Goulding, M.D.; Chalepakis, G.; Deutsch, U.; Erselius, J.R.; Gruss, P. Pax-3, a novel murine DNA binding protein expressed during early neurogenesis. EMBO J. 1991, 10, 1135–1147. [Google Scholar] [CrossRef]
- McFadden, A.; Martin, K.; Vierra, M.; Robilliard, H.; Lundquist, E.W.; Everts, R.E.; Brooks, S.A.; Lafayette, C. Three HPS5 Mutations Associated with Depigmentation in Diverse Horse Breeds. J. Livest. Sci. 2023; Manuscript submitted for review. [Google Scholar]
- Santschi, E.M.; Vrotsos, P.D.; Purdy, A.K.; Mickelson, J.R. Incidence of the endothelin receptor B mutation that causes lethal white foal syndrome in white-patterned horses. Am. J. Vet. Res. 2001, 62, 97–103. [Google Scholar] [CrossRef]
- Santschi, E.M.; Purdy, A.K.; Valberg, S.J.; Vrotsos, P.D.; Kaese, H.; Mickelson, J.R. Endothelin receptor B polymorphism associated with lethal white foal syndrome in horses. Mamm. Genome 1998, 9, 306–309. [Google Scholar] [CrossRef] [PubMed]
- Holl, H.M.; Brooks, S.A.; Archer, S.; Brown, K.; Malvick, J.; Penedo, M.C.T.; Bellone, R.R. Variant in the RFWD3 gene associated with PATN1, a modifier of leopard complex spotting. Anim. Genet. 2016, 47, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Rockwell, H.; Mack, M.; Famula, T.; Sandmeyer, L.; Bauer, B.; Dwyer, A.; Lassaline, M.; Beeson, S.; Archer, S.; McCue, M.; et al. Genetic investigation of equine recurrent uveitis in Appaloosa horses. Anim. Genet. 2020, 51, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Sandmeyer, L.S.; Kingsley, N.B.; Walder, C.; Archer, S.; Leis, M.L.; Bellone, R.R.; Bauer, B.S. Risk factors for equine recurrent uveitis in a population of Appaloosa horses in western Canada. Vet. Ophthalmol. 2020, 23, 515–525. [Google Scholar] [CrossRef] [PubMed]
- Esdaile, E.S. Short Tandem Repeat Analysis of Genetic Diversity Metrics in American Standardbreds and an Investigation on the Cause of the Rabicano Coat Color Phenotype Utilizing Short and Long Read Sequencing Methods. [UC Davis]. 2021. Available online: https://escholarship.org/uc/item/95w0q51r (accessed on 17 July 2023).


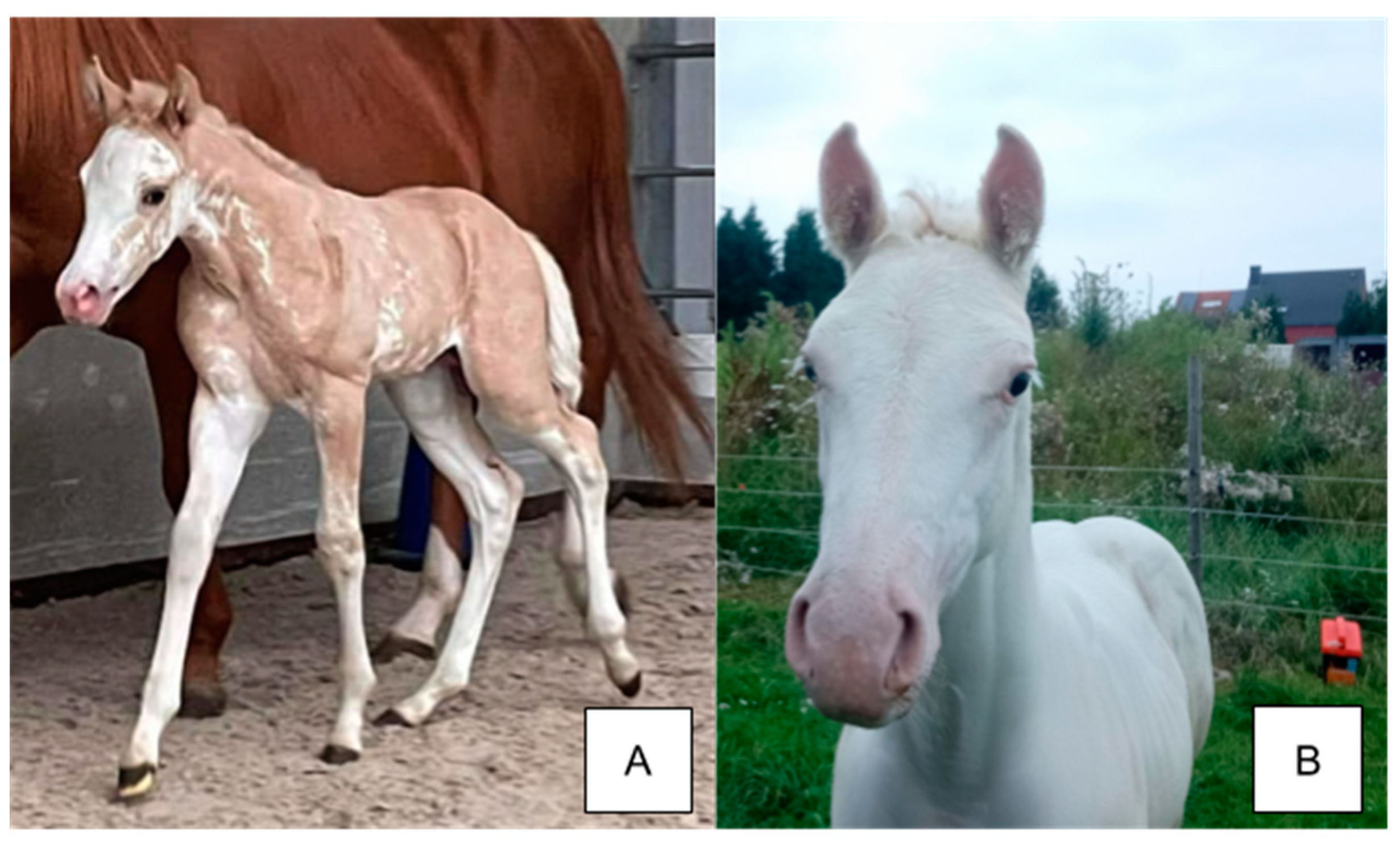

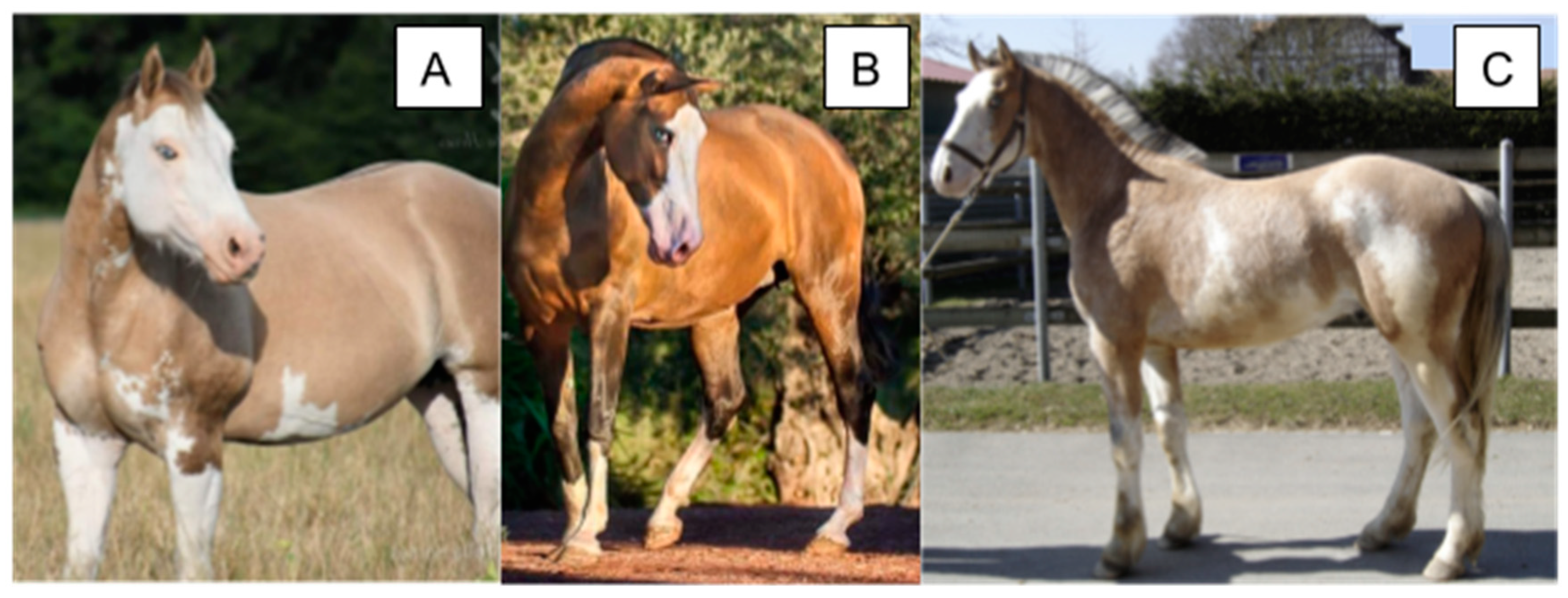


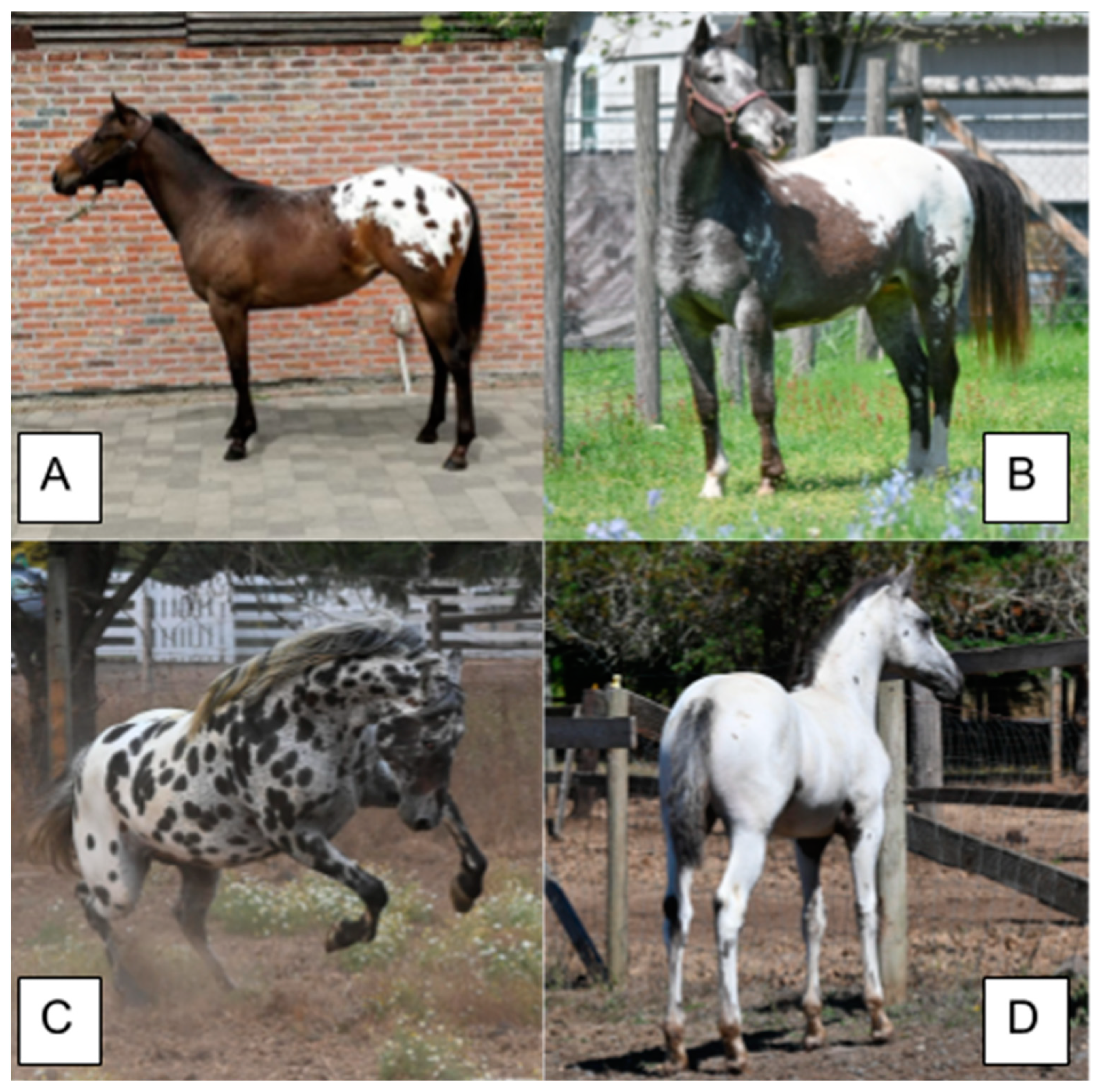
| Trait | Allele Symbol | Gene/Region | Encoded Protein (Complex) | Distinguishing Characteristics (Phenotype) |
|---|---|---|---|---|
| Dominant White | W | KIT | KIT | White spots or white coat, pink skin (sabino-like, all white) |
| Tobiano | TO | KIT | KIT | Large white spots covering the body and crossing the spine, white legs, pink skin (tobiano) |
| Sabino | SB1 | KIT | KIT | Jagged white markings or white coat, pink skin (sabino, all white) |
| Roan | RN | KIT | KIT | Interspersed white hairs distributed through the coat (roan) |
| Splashed White | SW | MITF, PAX3 | MITF, PAX3 | Deafness, blue eyes, smooth boarded white faces, abdomens, and legs (splashed white) |
| Eden White | EDXW | HPS5 | HPS5 (BLOC-2) | White body spots, legs, and faces (sabino-like) |
| Lethal White Overo | LWO, O | EDNRB | EDNRB | White face, legs, and body spots that do not cross the spine (overo) |
| Leopard Spotting | LP | TRPM1 | TRPM1 | White hind quarter or white coat with colored spots, epistatic to PATN1 (blanket or leopard) |
| Pattern 1 | PATN1 | RFWD3 | RFWD3 | Increases white spotting for individuals with a least one copy of LP |
| Allele | Genomic Coordinate EquCab3.0 | Type | Phenotype | Homozygotes | References |
|---|---|---|---|---|---|
| W1 | chr3:79545942G>C | nonsense | All White | Not Observed | [2] |
| W2 | chr3:79549540C>T | missense | All White | Not Observed | [2] |
| W3 | chr3:79578535T>A | nonsense | All White | Not Observed | [2] |
| W4 | chr3:79549780G>A | missense | All White | Not Observed | [2] |
| W5 | chr3:79545900delC | small deletion | Sabino-like | Not Observed | [12] |
| W6 | chr3:79573754C>T | missense | Sabino-like to All White | Not Observed | [12] |
| W7 | chr3:79580000C>G | splice site | All White | Not Observed | [12] |
| W8 | chr3:79545374C>T | splice site | Sabino-like | Not Observed | [12] |
| W9 | chr3:79549797C>T | missense | All White | Not Observed | [12] |
| W10 | chr3:79566925_79566928del | small deletion | Sabino-like to All White | Not Observed | [12] |
| W11 | chr3:79540429C>A | splice site | All White | Not Observed | [12] |
| W12 | chr3:79579755_79579779delAGACG | small deletion | Sabino-like | Not Observed | [17] |
| W13 | chr3:79544066C>G | splice site | All White | Not Observed | [14] |
| W14 | chr3:79544151_79544204del | gross deletion | All White | Not Observed | [14] |
| W15 | chr3:79550351A>G | missense | Sabino-like to All White | Observed | [14,18] |
| W16 | chr3:79540741T>A | missense | All White | Not Observed | [14] |
| W17a | chr3:79548265T>A | missense | All White | Not Observed | [14] |
| W17b | chr3:79548244A>G | missense | All White | Not Observed | [14] |
| W18 | chr3:79553751C>T | splice site | Sabino-like | Not Observed | [15] |
| W19 | chr3:79553776T>C | missense | Sabino-like | Observed | [15] |
| W20 | chr3:7948220T>C | missense | No markings to Sabino-like | Observed | [15] |
| W21 | chr3:79544174delG | small deletion | Sabino-like | Not Observed | [13] |
| W22 | chr3:79548925_79550822del1898 | gross deletion | Sabino-like | Not Observed | [11] |
| W23 | chr3:79578484C>G | splice site | All White | Not Observed | [18] |
| W24 | chr3:79545245C>T | splice site | All White | Not Observed | [10] |
| W25 | chr3:77769878T>C | missense | All White | Not Observed | [16] |
| W26 | chr3:79544150del | small deletion | Sabino-like | Not Observed | [16] |
| W27 | chr3:79552028A>C | missense | All White | Not Observed | [16] |
| W28 | chr3:79579925_79581197del | gross deletion | Sabino-like | Not Observed | [19] |
| W30 | chr3:79548244T>A | missense | All White | Not Observed | [20] |
| W31 | chr3:79618532_79618533insT | fs nonsense | Sabino-like | Not Observed | [7] |
| W32 | chr3:79538738C>T | missense | No markings to Sabino-like | Observed | [7] |
| W33 | chr3:79545248T>A | missense | Sabino-like | Not Observed | [5] |
| W34 | chr3:79566881T>C | missense | No markings to Sabino-like | Observed | [8] |
| W35 | chr3:79618649A>C | UTR variant | No markings to Sabino-like | Observed | [6] |
| Allele | Gene | Genome Coordinate EquCab3.0 | Type | Phenotype | Reference |
|---|---|---|---|---|---|
| SB1 | KIT | chr3:79544206A>T | splice site | White markings with jagged borders and pink skin. Homozygotes are all white with pink skin | [9] |
| TO | Intergenic | chr3:42737476_79471993inv | paracentric inversion | Large white areas across the body, pink skin | [36] |
| RN | KIT | chr3:79543439A>G | associated SNP | White hairs distributed throughout the coat | [37] |
| Allele | Gene | Genomic Coordinate EquCab3.0 | Type | Phenotype | Reference |
|---|---|---|---|---|---|
| SW1 | MITF | chr16:21579201delinsATAATAACCTA | small indel | Splashed Markings, Deafness, Blue Eyes | [44] |
| SW2 | PAX3 | chr6:11199026C>T | missense | Minimal Splashed Markings, Deafness, Blue Eyes | [44] |
| SW3 | MITF | chr16:21567245_21567249del | small deletion | Splashed Markings, Deafness, Blue Eyes | [44] |
| SW4 | PAX3 | chr6:11199140G>C | missense | Minimal Splashed Markings, Blue Eyes | [15] |
| SW5 | MITF | chr16:21503211_21566617del | gross deletion | Splashed Markings, Deafness, Blue Eyes | [45] |
| SW6 | MITF | chr16:21551060_21559770del | gross deletion | Splashed Markings, Deafness, Blue Eyes | [46] |
| SW7 | MITF | chr16:21559953_21559955del | small deletion | Splashed Markings, Deafness, Blue Eyes | [48] |
| SW8 | MITF | chr16:21555811_21558139del | gross deletion | Splashed Markings, Deafness, Blue Eyes | [43] |
| SW9 | MITF | chr16:21559940A>T | missense | Splashed Markings, Blue Eyes | [47] |
| SW10 | PAX3 | chr6:11196181C>T | nonsense | Splashed Markings, Blue Eyes | [47] |
| Allele | Genomic Coordinate EquCab3.0 | Type | Phenotype | Reference |
|---|---|---|---|---|
| EDXW1 | chr7:88741616T>A | missense | High socks, belly spots, blazes | [56] |
| EDXW2 | chr7:88751667G>A | missense | High socks, belly spots, blazes | [56] |
| EDXW3 | chr7:88766673T>C | splice site | High socks, belly spots, blazes | [56] |
| Allele | Gene | Genomic Coordinate EquCab3.0 | Type | Phenotype | Reference |
|---|---|---|---|---|---|
| LP | TRPM1 | chr1:108297929_108297930insN[1378] | insertion | White hind quarter, all white (hom), leopard spots | [27] |
| PATN1 | RFWD3 | chr3:24352525T>G | UTR variant | Increases white and decreases number of spots only for LP/LP or LP/lp individuals | [59] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
McFadden, A.; Vierra, M.; Martin, K.; Brooks, S.A.; Everts, R.E.; Lafayette, C. Spotting the Pattern: A Review on White Coat Color in the Domestic Horse. Animals 2024, 14, 451. https://doi.org/10.3390/ani14030451
McFadden A, Vierra M, Martin K, Brooks SA, Everts RE, Lafayette C. Spotting the Pattern: A Review on White Coat Color in the Domestic Horse. Animals. 2024; 14(3):451. https://doi.org/10.3390/ani14030451
Chicago/Turabian StyleMcFadden, Aiden, Micaela Vierra, Katie Martin, Samantha A. Brooks, Robin E. Everts, and Christa Lafayette. 2024. "Spotting the Pattern: A Review on White Coat Color in the Domestic Horse" Animals 14, no. 3: 451. https://doi.org/10.3390/ani14030451








