Pedigree-Based Genetic Diversity in the South African Boerboel Dog Breed
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Pedigree Data
2.2. Pedigree Analysis
2.2.1. Pedigree Completeness
2.2.2. Estimation of the Inbreeding Coefficient
2.2.3. Estimation of the Probabilities of Gene Origin
3. Results
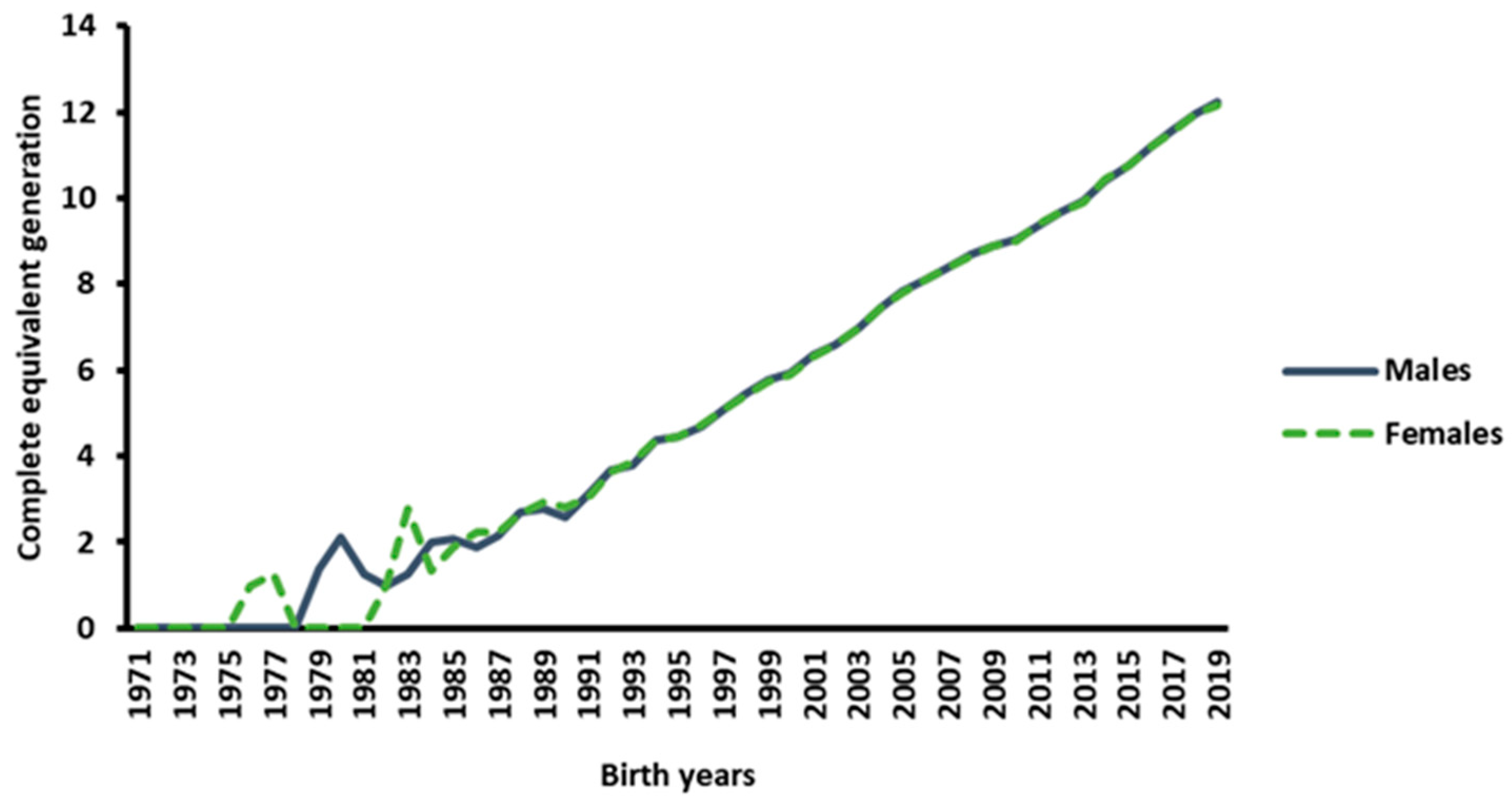
3.1. Pedigree Depth and Completeness
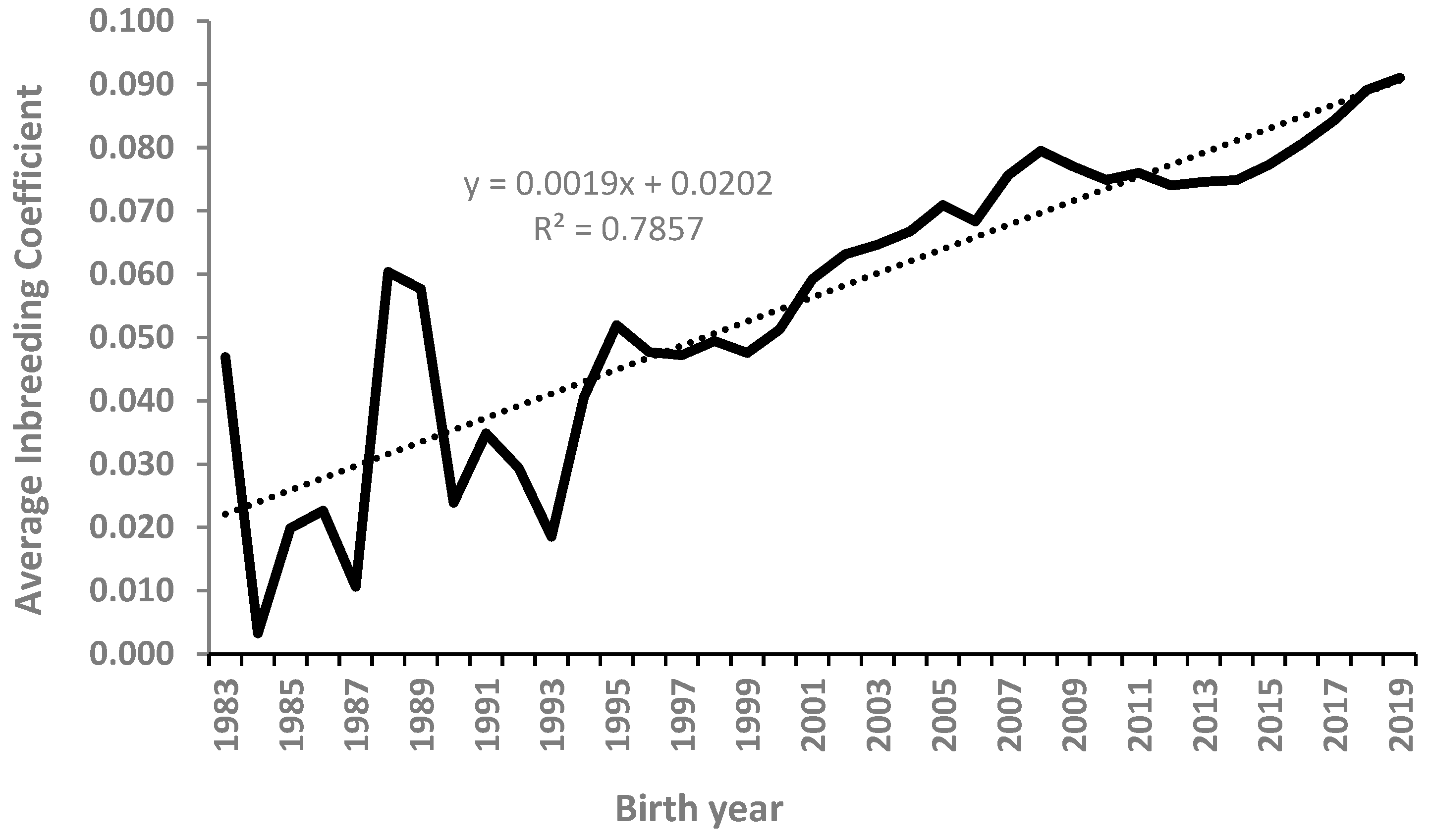
3.2. Estimated Inbreeding Coefficients
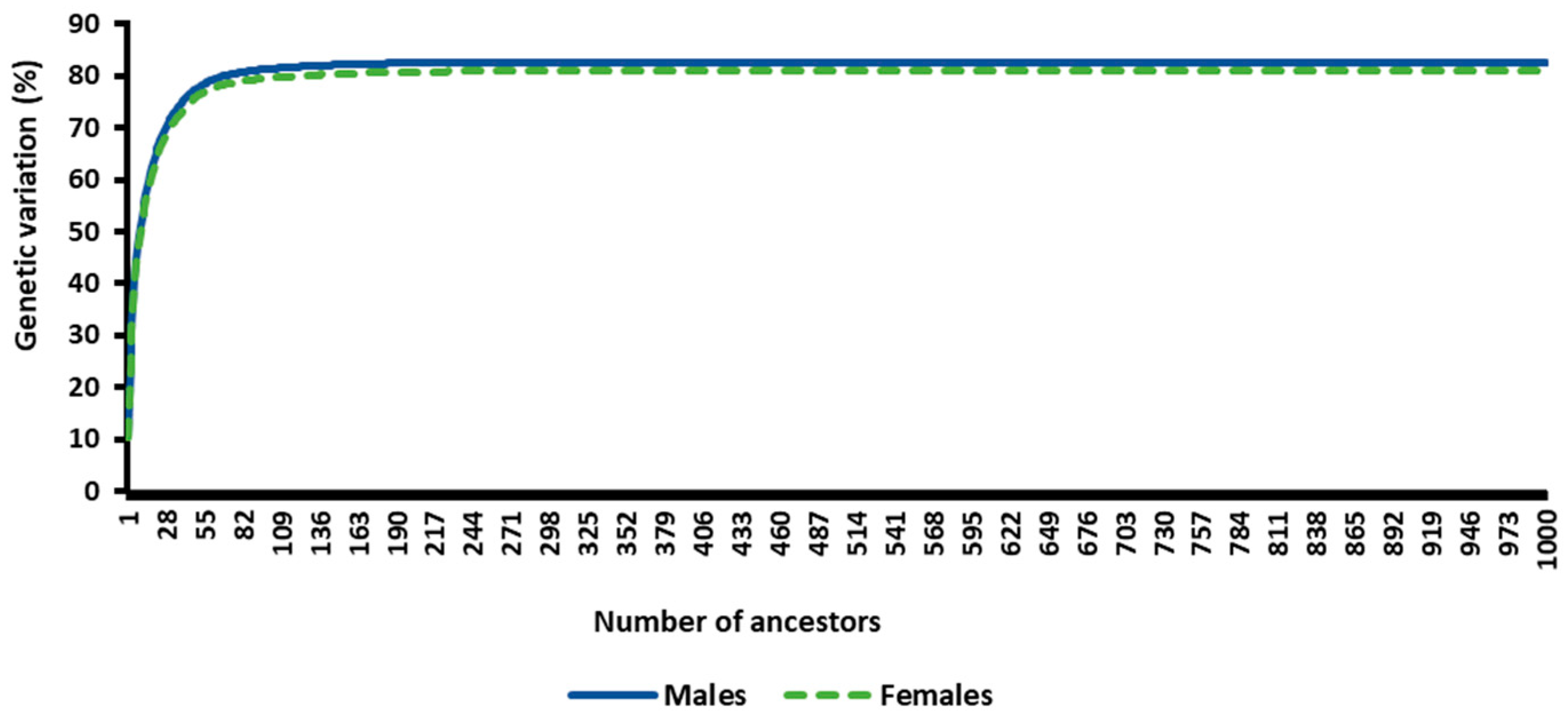
3.3. Estimated Probabilities of Gene Origin
Estimated Measures of Genetic Diversity
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Swart, S. Dogs and dogma: A discussion of the socio-political construction of Southern African dog ‘breeds’ as a window onto social history. In Canis Africanis; Brill: Leiden, The Netherlands, 2008; pp. 267–287. [Google Scholar]
- Van Sittert, L.; Swart, S. Canis familiaris: A dog history of Southern Africa. In Canis Africanis; Brill: Leiden, The Netherlands, 2008; pp. 1–34. [Google Scholar]
- Van Sittert, L.; Swart, S. Canis familiaris: A dog history of South Africa. S. Afr. Hist. J. 2003, 48, 138–173. [Google Scholar] [CrossRef]
- South African Boerboel Breeders’ Society (SABBS). Boerboel Breed Standard. 2019. Available online: https://sabbs.org/the-boerboel/breed-standard (accessed on 25 April 2021).
- Kennel Union of Southern Africa (KUSA). Breed Standard. 2008. Available online: https://www.kusa.co.za/index.php/documents/breed-standards/working-group-list (accessed on 2 February 2023).
- Theron, H. Genetic diversity in the Boerboel breed. South African Studbook. Boerboel J. 2019, 9–13. Available online: https://www.sabbs.co.za/forms/2de28d39-2fa6-4fa9-afb1-35f9836d0755 (accessed on 3 March 2022).
- Mastrangelo, S.; Biscarini, F.; Tolone, M.; Auzino, B.; Ragatzu, M.; Spaterna, A.; Ciampolini, R. Genomic characterization of the Braque Français type Pyrénées dog and relationship with other breeds. PLoS ONE 2018, 13, e0208548. [Google Scholar] [CrossRef]
- Mäki, K. Population structure and genetic diversity of worldwide Nova Scotia Duck Tolling Retriever and Lancashire Heeler dog populations. J. Anim. Breed. Genet. 2010, 127, 318–326. [Google Scholar] [CrossRef]
- Martínez, M.E.; Calderón, C.; Uribe, H.; De la Barra, R. Effect of management practices in the productive performance of three sheep breeds in the Chiloé Archipelago, Chile. J. Livest. Sci. 2012, 3, 57–66. [Google Scholar]
- Windig, J.J.; Hulsegge, I. Retriever and pointer: Software to evaluate inbreeding and genetic management in captive populations. Animals 2021, 11, 1332. [Google Scholar] [CrossRef] [PubMed]
- Sattaei Mokhtari, M.; Miraei-Ashtiani, S.R.; Jafaroghli, M.; Gutiérrez, J.P. Studying genetic diversity in Moghani sheep using pedigree analysis. J. Agric. Sci. Technol. 2015, 17, 1151–1160. [Google Scholar]
- Stachowicz, K.; Brito, L.F.; Oliveira, H.R.; Miller, S.P.; Schenkel, F.S. Assessing genetic diversity of various Canadian sheep breeds through pedigree analyses. Can. J. Anim. Sci. 2018, 98, 741–749. [Google Scholar] [CrossRef]
- Ólafsdóttir, G.Á.; Kristjánsson, T. Correlated pedigree and molecular estimates of inbreeding and their ability to detect inbreeding depression in the Icelandic sheepdog, a recently bottlenecked population of domestic dogs. Conserv. Genet. 2008, 9, 1639–1641. [Google Scholar] [CrossRef]
- Jansson, M. Assessing Inbreeding and Loss of Genetic Variation in Canids, Domestic Dog (Canis familiaris) and Wolf (Canis lupus), Using Pedigree Data. Ph.D. Thesis, Stockholm University, Stockholm, Sweden, 2014. [Google Scholar]
- Vostrá-Vydrová, H.; Vostrý, L.; Hofmanová, B.; Krupa, E.; Zavadilová, L. Pedigree analysis of the endangered Old Kladruber horse population. Livest. Sci. 2016, 185, 17–23. [Google Scholar] [CrossRef]
- Núñez-Domínguez, R.; Martínez-Rocha, R.E.; Hidalgo-Moreno, J.A.; Ramírez-Valverde, R.; García-Muñiz, J.G. Evaluation of the Romosinuano cattle population structure in Mexico using pedigree analysis. Rev. Colomb. Cienc. Pecu. 2020, 33, 44–59. [Google Scholar] [CrossRef]
- Toro, A.; Mahfouz, A.E.; Ardiri, A.; Malaguarnera, M.; Malaguarnera, G.; Loria, F.; Bertino, G.; Di Carlo, I. What is changing in indications and treatment of hepatic hemangiomas. A review. Ann. Hepatol. 2014, 13, 327–339. [Google Scholar] [CrossRef]
- Addo, S.; Schäler, J.; Hinrichs, D.; Thaller, G. Genetic diversity and ancestral history of the German Angler and the Red-and-White dual-purpose cattle breeds assessed through pedigree analysis. Agric. Sci. 2017, 8, 1033–1047. [Google Scholar] [CrossRef][Green Version]
- Eusebi, P.G.; Martinez, A.; Cortes, O. Genomic tools for effective conservation of livestock breed diversity. Diversity 2019, 12, 8. [Google Scholar] [CrossRef]
- Leroy, G. Genetic diversity, inbreeding and breeding practices in dogs: Results from pedigree analyses. Vet. J. 2011, 189, 177–182. [Google Scholar] [CrossRef]
- Gajaweera, C.; Kang, J.M.; Lee, D.H.; Lee, S.H.; Kim, Y.K.; Wijayananda, H.I.; Kim, J.J.; Ha, J.H.; Choi, B.H.; Lee, S.H. Genetic diversity and population structure of the Sapsaree, a native Korean dog breed. BMC Genet. 2019, 20, 66. [Google Scholar] [CrossRef]
- Radko, A.; Podbielska, A. Microsatellite DNA analysis of genetic diversity and parentage testing in the popular dog breeds in Poland. Genes 2021, 12, 485. [Google Scholar] [CrossRef]
- González-Cano, R.; González-Martínez, A.; Muñoz-Mejías, M.E.; Valera, P.; Rodero, E. Analyses of Genetic Diversity in the Endangered “Berrenda” Spanish Cattle Breeds Using Pedigree Data. Animals 2022, 12, 249. [Google Scholar] [CrossRef] [PubMed]
- Boichard, D. Pedig: A fortran package for pedigree analysis suited for large populations. In Proceedings of the 7th World Congress on Genetics Applied to Livestock Production, Institut National de la Recherche Agronomique (INRA), Montpellier, France, 19–23 August 2002. [Google Scholar]
- Boichard, D.; Maignel, L.; Verrier, E. The value of using probabilities of gene origin to measure genetic variability in a population. Genet. Sel. Evol. 1997, 29, 5–23. [Google Scholar] [CrossRef]
- Vigh, Z.; Gyovai, P.; Csato, L.; Bokor, A.; Farkas, J.; Radnoczi, L.; Komlosi, I.; Nagy, I. Effect of inbreeding on lean meat percentage and average daily gain in Hungarian Landrace pigs. Arch. Anim. Breed. 2008, 51, 541–548. [Google Scholar] [CrossRef]
- Meuwissen, T.I.; Luo, Z. Computing coefficient of consanguinitys in large populations. Genet. Sel. Evol. 1992, 24, 305–313. [Google Scholar] [CrossRef]
- Mc Parland, S.; Kearney, J.F.; Rath, M.; Berry, D.P. Inbreeding trends and pedigree analysis of Irish dairy and beef cattle populations. J. Anim. Sci. 2007, 85, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Weigel, K.A. Controlling inbreeding in modern breeding programs. J. Dairy Sci. 2001, 84, E177–E184. [Google Scholar] [CrossRef]
- Mostert, B.E.; van Marle-Köster, E.; Visser, C.; Oosthuizen, M. Genetic analysis of pre-weaning survival and inbreeding in the Boxer dog breed of South Africa. S. Afr. J. Anim. Sci. 2015, 45, 476–484. [Google Scholar] [CrossRef][Green Version]
- Food and Agriculture Organization. Initiative for Domestic Animal Diversity. Secondary Guidelines for Development of National Farm Animal Genetic Resources Management Plans: Management of Small Populations at Risk; FAO: Rome, Italy, 1998. [Google Scholar]
- Meuwissen, T.H.E. Maximizing the response of selection with a predefined rate of inbreeding. J. Anim. Sci. 1997, 75, 934–940. [Google Scholar] [CrossRef]
- Sørensen, A.C.; Sørensen, M.K.; Berg, P. Inbreeding in Danish dairy cattle breeds. J. Dairy Sci. 2005, 88, 1865–1872. [Google Scholar] [CrossRef]
- Shariflou, M.R.; James, J.W.; Nicholas, F.W.; Wade, C.M. A genealogical survey of Australian registered dog breeds. Vet. J. 2011, 189, 203–210. [Google Scholar] [CrossRef]
- Machová, K.; Kranjčevičová, A.; Vostrý, L.; Krupa, E. Analysis of Genetic Diversity in the Czech Spotted Dog. Animals 2020, 10, 1416. [Google Scholar] [CrossRef]
- Food and Agriculture Organization. In Vivo Conservation of Animal Genetic Resources. In FAO Animal Production and Health Guidelines; FAO: Rome, Italy, 2013. [Google Scholar]
- Li, M.H.; Strandén, I.; Kantanen, J. Genetic diversity and pedigree analysis of the Finnsheep breed. J. Anim. Sci. 2009, 87, 1598–1605. [Google Scholar] [CrossRef] [PubMed]
- Yepes, W.; Pardo Pérez, E.; Causil Vargas, L.A. Genetic Diversity of the Creole Horse (Equus caballus) through Genes Associated with The Coat in Valencia, Colombia. Rev. Investig. Vet. Perú 2017, 28, 562–570. [Google Scholar] [CrossRef][Green Version]
- Ács, V.; Bokor, Á.; Nagy, I. Population structure analysis of the border collie dog breed in Hungary. Animals 2019, 9, 250. [Google Scholar] [CrossRef] [PubMed]
- Navas, C.M.; González, F.J.N.; López, V.C.; Capellà, L.P.; Fernández, M.G.; Bermejo, J.V.D. Impact of breeding for coat and spotting patterns on the population structure and genetic diversity of an islander endangered dog breed. Res. Vet. Sci. 2020, 131, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Carolino, N.; Gama, L.T. Indicators of genetic erosion in an endangered population: The Alentejana cattle breed in Portugal. J. Anim. Sci. 2008, 86, 47–56. [Google Scholar] [CrossRef]
- Krupa, E.; Žáková, E.; Krupová, Z. Evaluation of inbreeding and genetic variability of five pig breeds in Czech Republic. Asian-Australas. J. Anim. Sci. 2015, 28, 25. [Google Scholar] [CrossRef]
- Kania-Gierdziewicz, J.; Gierdziewicz, M.; Budzynski, B. Genetic structure analysis of Tatra Shepherd dog population from Tatra Mountain region. Ann. Anim. Sci. 2015, 15, 323–335. [Google Scholar] [CrossRef]
- Cecchi, F.; Paci, G.; Spaterna, A.; Ciampolini, R. Genetic variability in Bracco Italiano dog breed assessed by pedigree data. Ital. J. Anim. Sci. 2013, 12, e54. [Google Scholar] [CrossRef]
- Nielen, A.L.J.; Van Der Beek, S.; Ubbink, G.J.; Knol, B.W. Epidemiology: Population parameters to compare dog breeds: Differences between five dutch purebred populations. Vet. Q. 2001, 23, 43–49. [Google Scholar] [CrossRef]
- Leroy, G.; Rognon, X.; Varlet, A.; Joffrin, C.; Verrier, E. Genetic variability in French dog breeds assessed by pedigree data. J. Anim. Breed. Genet. 2006, 123, 1–9. [Google Scholar] [CrossRef]
- Pekkala, N.; Knott, K.E.; Kotiaho, J.S.; Nissinen, K.; Puurtinen, M. The effect of inbreeding rate on fitness, inbreeding depression and heterosis over a range of inbreeding coefficients. Evol. Appl. 2014, 7, 1107–1119. [Google Scholar] [CrossRef]
- Wijnrocx, K.; François, L.; Stinckens, A.; Janssens, S.; Buys, N. Half of 23 Belgian dog breeds has a compromised genetic diversity, as revealed by genealogical and molecular data analysis. J. Anim. Breed. Genet. 2016, 133, 375–383. [Google Scholar] [CrossRef]
- Meuwissen, T.A.; Woolliams, J.A. Effective sizes of livestock populations to prevent a decline in fitness. Theor. Appl. Genet. 1994, 89, 1019–1026. [Google Scholar] [CrossRef]
- Willi, Y.; Kristensen, T.N.; Sgrò, C.M.; Weeks, A.R.; Ørsted, M.; Hoffmann, A.A. Conservation genetics as a management tool: The five best-supported paradigms to assist the management of threatened species. Proc. Natl. Acad. Sci. USA 2022, 119, e2105076119. [Google Scholar] [CrossRef]
- Makina, S.O.; Muchadeyi, F.C.; van Marle-Köster, E.; MacNeil, M.D.; Maiwashe, A. Genetic diversity and population structure among six cattle breeds in South Africa using a whole genome SNP panel. Front. Genet. 2014, 5, 333. [Google Scholar] [CrossRef]
- Fernandes, S.D.; Malovrh, Š.; Kovac, M.; Cadavez, V. Study of genetic diversity of Bísaro pigs breed by pedigree analysis. Lucr. Ştiinţifice Ser. Zooteh. 2010, 178, 326–330. [Google Scholar]
- Ocampo Gallego, R.; Ramírez Toro, J.; Lopera Peña, J.; Restrepo Castañeda, G.; Gallego Gil, J. Genetic diversity assessed by pedigree analysis in the Blanco Orejinegro (BON) cattle breed population from the Colombian germplasm bank. Chil. J. Agric. Anim. Sci. 2020, 36, 69–77. [Google Scholar] [CrossRef]
- Pérez-González, J.; Costa, V.; Santos, P.; Slate, J.; Carranza, J.; Fernández-Llario, P.; Zsolnai, A.; Monteiro, N.M.; Anton, I.; Buzgó, J.; et al. Males and females contribute unequally to offspring genetic diversity in the polygynandrous mating system of wild boar. PLoS ONE 2014, 9, e115394. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Barros, E.A.; Brasil, L.D.A.; Tejero, J.P.; Delgado-Bermejo, J.V.; Ribeiro, M.N. Population structure and genetic variability of the Segureña sheep breed through pedigree analysis and inbreeding effects on growth traits. Small Rumin. Res. 2017, 149, 128–133. [Google Scholar] [CrossRef]
- Stankowski, S. Ecological speciation in an island snail: Evidence for the parallel evolution of a novel ecotype and maintenance by ecologically dependent postzygotic isolation. Mol. Ecol. 2013, 22, 2726–2741. [Google Scholar] [CrossRef] [PubMed]
- Michels, P.W.; Distl, O. Genetic Diversity and Trends of Ancestral and New Inbreeding in Deutsch Drahthaar Assessed by Pedigree Data. Animals 2022, 12, 929. [Google Scholar] [CrossRef] [PubMed]
- Brito, F.V.; Sargolzaei, M.; Braccini Neto, J.; Cobuci, J.A.; Pimentel, C.M.; Barcellos, J.; Schenkel, F.S. In-depth pedigree analysis in a large Brazilian Nellore herd. Genet. Mol. Res. 2013, 12, 5758–5765. [Google Scholar] [PubMed]
- Sölkner, J.; Filipcic, L.; Hampshire, N. Genetic variability of populations and similarity of subpopulations in Austrian cattle breeds determined by analysis of pedigrees. Anim. Sci. 1998, 67, 249–256. [Google Scholar] [CrossRef]
- Domínguez-Viveros, J.; Reyes-Cerón, A.; Saiz-Pineda, J.F.; Villegas-Gutiérrez, C.; Aguilar-Palma, G.N.; Rodríguez-Almeida, F.A. Structure and genetic variability of the Mexican Sardo Negro breed. Ciência Rural 2021, 52, 5. [Google Scholar] [CrossRef]
- Fernández, J.; Meuwissen, T.H.E.; Toro, M.A.; Mäki-Tanila, A. Management of genetic diversity in small farm animal populations. Animal 2011, 5, 1684–1698. [Google Scholar] [CrossRef]
- Lampi, S.; Donner, J.; Anderson, H.; Pohjoismäki, J. Variation in breeding practices and geographic isolation drive subpopulation differentiation, contributing to the loss of genetic diversity within dog breed lineages. Canine Med. Genet. 2020, 7, 5. [Google Scholar] [CrossRef]
- Maignel, L.; Boichard, D.; Verrier, E. Genetic variability of French dairy breeds estimated from pedigree information. INTERBULL Bull. 1996, 14, 49. [Google Scholar]
- Carolino, N.; Vitorino, A.; Carolino, I.; Pais, J.; Henriques, N.; Silveira, M.; Vicente, A. Genetic diversity in the Portuguese mertolenga cattle breed assessed by pedigree analysis. Animals 2020, 10, 1990. [Google Scholar] [CrossRef] [PubMed]
- Voges, S.; Distl, O. Inbreeding trends and pedigree analysis of Bavarian Mountain hounds, Hanoverian hounds and Tyrolean hounds. J. Anim. Breed. Genet. 2009, 126, 357–365. [Google Scholar] [CrossRef]
- Dreger, D.L.; Rimbault, M.; Davis, B.W.; Bhatnagar, A.; Parker, H.G.; Ostrander, E.A. Whole-genome sequence, SNP chips and pedigree structure: Building demographic profiles in domestic dog breeds to optimize genetic-trait mapping. Dis. Models Mech. 2016, 9, 1445–1460. [Google Scholar] [CrossRef]
- Soh, P.X.Y.; Hsu, W.T.; Khatkar, M.S.; Williamson, P. Evaluation of genetic diversity and management of disease in Border Collie dogs. Sci. Rep. 2021, 11, 6243. [Google Scholar] [CrossRef] [PubMed]
- Bannasch, D.L.; Kaelin, C.B.; Letko, A.; Loechel, R.; Hug, P.; Jagannathan, V.; Henkel, J.; Roosje, P.; Hytönen, M.K.; Lohi, H.; et al. Dog colour patterns explained by modular promoters of ancient canid origin. Nat. Ecol. Evol. 2021, 5, 1415–1423. [Google Scholar] [CrossRef]
- Kadlec, T.; Vrba, P.; Kepka, P.; Schmitt, T.; Konvicka, M. Tracking the decline of the once-common butterfly: Delayed oviposition, demography and population genetics in the hermit Chazara briseis. Anim. Conserv. 2010, 13, 172–183. [Google Scholar] [CrossRef]
- Peacock, M.M.; Dochtermann, N.A. Evolutionary potential but not extinction risk of Lahontan cutthroat trout (Oncorhynchus clarkii henshawi) is associated with stream characteristics. Can. J. Fish. Aquat. Sci. 2012, 69, 615–626. [Google Scholar] [CrossRef]
- Bozzi, R.; Franci, O.; Forabosco, F.; Pugliese, C.; Crovetti, A.; Filippini, F. Genetic variability in three Italian beef cattle breeds derived from pedigree information. Ital. J. Anim. Sci. 2006, 5, 129–137. [Google Scholar] [CrossRef]
- Valera, M.; Molina, A.; Gutiérrez, J.P.; Gómez, J.; Goyache, F. Pedigree analysis in the Andalusian horse: Population structure, genetic variability and influence of the Carthusian strain. Livest. Prod. Sci. 2005, 95, 57–66. [Google Scholar] [CrossRef]
| Inbreeding Class | No. of Individuals | Proportion of Animals% |
|---|---|---|
| 0–5 | 28,664 | 35.8 |
| 5–10 | 34,668 | 43.3 |
| 10–15 | 9538 | 11.9 |
| 15–20 | 4396 | 5.5 |
| 20–25 | 1225 | 1.5 |
| 25–30 | 1067 | 1.3 |
| 30–35 | 340 | 0.4 |
| 35–40 | 100 | 0.1 |
| 40–45 | 15 | 0.02 |
| 50–55 | 1 | 0 |
| Number of individuals: 87,755 | ||
| Number of inbred individuals: 80,014 | ||
| Mean inbreeding of inbred individuals: 7.5 | ||
| Maximum inbreeding: 50 | ||
| Parameters | Males | Females |
|---|---|---|
| Total no. of founders () | 348 | 356 |
| Effective no. of founders () | 57.4 | 60.1 |
| Effective no. of ancestors () | 27.93 | 29.5 |
| f | 0.16 | 0.17 |
| 2.1 | 2.0 | |
| Effective population size () | 83.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mabunda, R.S.; Nephawe, K.A.; Mtileni, B.; Makgahlela, M.L. Pedigree-Based Genetic Diversity in the South African Boerboel Dog Breed. Animals 2024, 14, 975. https://doi.org/10.3390/ani14060975
Mabunda RS, Nephawe KA, Mtileni B, Makgahlela ML. Pedigree-Based Genetic Diversity in the South African Boerboel Dog Breed. Animals. 2024; 14(6):975. https://doi.org/10.3390/ani14060975
Chicago/Turabian StyleMabunda, Ripfumelo Success, Khathutshelo Agree Nephawe, Bohani Mtileni, and Mahlako Linah Makgahlela. 2024. "Pedigree-Based Genetic Diversity in the South African Boerboel Dog Breed" Animals 14, no. 6: 975. https://doi.org/10.3390/ani14060975
APA StyleMabunda, R. S., Nephawe, K. A., Mtileni, B., & Makgahlela, M. L. (2024). Pedigree-Based Genetic Diversity in the South African Boerboel Dog Breed. Animals, 14(6), 975. https://doi.org/10.3390/ani14060975







