Effect of SNPs in the Promoter Region on the Expression of Cytochrome P450 2E1 (CYP2E1) in Pig Liver
Abstract
:Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. SNP Discovery
2.2. Animals
2.3. Genotyping
2.4. Isolation of Total RNA
2.5. cDNA Synthesis and Real-Time PCR
2.6. Western Blotting
2.7. Statistical Analysis
3. Results
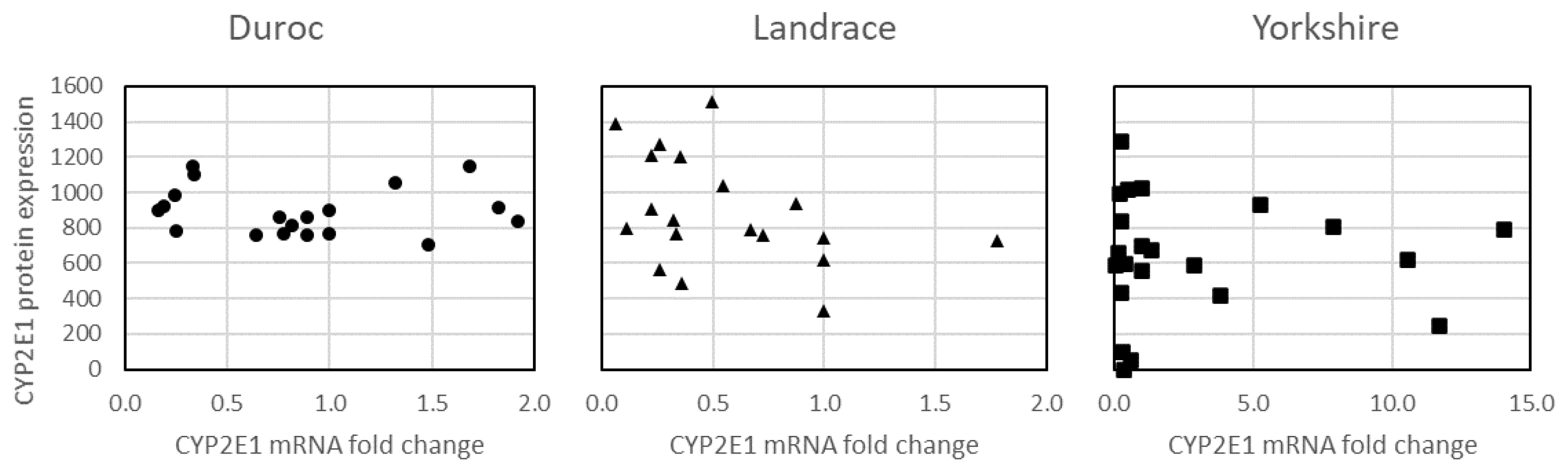
3.1. CYP2E1 mRNA and Protein Expression
3.2. SNP Effect on mRNA Expression
4. Discussion
4.1. Carcass Weight and CYP2E1 Expression
4.2. SNP Discovery and Effects on CYP2E1 mRNA and Protein Expression
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Patterson, R. 5α-androst-16-ene-3-one: Compound responsible for taint in boar fat. J. Sci. Food Agric. 1968, 19, 31–38. [Google Scholar] [CrossRef]
- Vold, E. Fleischproduktionseigenschaften bei Ebern und Kastraten. IV. Organoleptische und Gaschromatographische Untersuchungen Wasserdampfflüchtiger Stoffe des Rückenspecks von Ebern; Report No. 238; Institute of Animal Genetics and Breeding, NLH: Vollebekk, Norway, 1970. [Google Scholar]
- Squires, E.J.; Bone, C.; Cameron, J. Pork production with entire males: Directions for control of boar taint. Animals 2020, 10, 1665. [Google Scholar] [CrossRef] [PubMed]
- Duarte, D.A.S.; Schroyen, M.; Mota, R.R.; Vanderick, S.; Gengler, N. Recent genetic advances on boar taint reduction as an alternative to castration: A review. J. Appl. Genet. 2021, 62, 137–150. [Google Scholar] [CrossRef] [PubMed]
- Falany, N.C.; Comer, A.K.; Dooley, P.T.; Glatt, H. Human dehydroepiandrosterone sulfotransferase: Purification molecular cloning and characterization. Ann. N. Y. Acad. Sci. 1995, 774, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Gibson, G.; Skett, P. Introduction to Drug Metabolism; Nelson Thornes: Cheltenham, UK, 2001. [Google Scholar]
- Zamek-Gliszczynski, M.; Hoffmaster, K.; Nezasa, K.; Tallman, M.; Brouwer, K. Integration of hepatic drug transporters and phase II metabolizing enzymes: Mechanisms of hepatic excretion of sulfate, glucuronide, and glutathione metabolites. Eur. J. Pharm. Sci. 2006, 27, 447–486. [Google Scholar] [CrossRef]
- Squires, J.; Lundström, K. Relationship between cytochrome P450IIEI in liver and levels of skatole and its metabolites in entire male pigs. J. Anim. Sci. 1997, 75, 2506–2511. [Google Scholar] [CrossRef] [PubMed]
- Doran, E.; Whittington, F.; Wood, J.; McGivan, J. Cytochrome P450IIEI (CYP2E1) is induced by skatole and this induction is blocked by androstenone in isolated pig hepatocytes. Chem.-Biol. Interact. 2002, 140, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Whittington, F.; Nute, G.; Hughes, S.; McGivan, J.; Lean, I.; Wood, J.; Doran, E. Relationships between skatole and androstenone accumulation, and cytochrome P4502E1 expression in Meishan X Large White pigs. Meat Sci. 2004, 67, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Skinner, T.; Doran, E.; McGilvan, J.; Haley, C.; Archibald, A. Cloning and mapping of the porcine cytochrome-P450 2E1 gene and its association with skatole levels in the domestic pig. Anim. Genet. 2005, 36, 417–422. [Google Scholar] [CrossRef]
- Morlein, D.; Lungershausen, M.; Steinke, K.; Sharifi, A.; Knorr, C. A single nucleotide polymorphism in the CYP2E1 gene promoter affects skatole content in backfat of boars of two commercial Duroc-sired crossbred populations. Meat Sci. 2012, 92, 739–744. [Google Scholar] [CrossRef]
- Sargolzaei, M.; Chesnais, J.; Schenkel, F. A new approach for efficient genotype imputation using information from relatives. BMC Genom. 2014, 15, 478. [Google Scholar] [CrossRef] [PubMed]
- Gray, M.; Squires, J. Effects of nuclear receptor transactivation on steroid hormone synthesis and gene expression in porcine hepatocytes. J. Steroid Biochem. Mol. Biol. 2013, 133, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.; Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Billen, M.; Squires, E.J. The role of porcine cytochrome b5A and cytochrome b5B in the regulation of cytochrome P45017A1 activities. J. Steroid Biochem. Mol. Biol. 2009, 113, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Wiercinska, P.; Lou, Y.; Squires, E.J. The roles of different porcine cytochrome P450 enzymes and cytochrome b5A in skatole metabolism. Animal 2012, 6, 834–845. [Google Scholar] [CrossRef] [PubMed]
- Barhdadi, A.; Dubé, M.-P. Testing for Linkage Disequilibrium Using SAS/GENETICS and SAS/STAT; Statistical Genetics Research Group: Montreal, QC, Canada, 2011. [Google Scholar]
- Moe, M.; Lien, S.; Aasmundstad, T.; Meuwissen, T.H.E.; Hansen, M.H.S.; Bendixen, C.; Grindflek, E. Association between SNPs within candidate genes and compounds related to boar taint and reproduction. BMC Genet. 2009, 10, 32. [Google Scholar] [CrossRef] [PubMed]
- Kocarek, T.; Zangar, R.; Novak, R. Post-transcriptional regulation of rat CYP2E1 expression: Role of CYP2E1 mRNA untranslated regions in control of translational efficiency and message stability. Arch. Biochem. Biophys. 2000, 376, 180–190. [Google Scholar] [CrossRef] [PubMed]
- Zamaratskaia, G.; Chen, G.; Lundström, K. Effects of sex, weight, diet and hCG administration on levels of skatole and indole in the liver and hepatic activities of cytochromes P4502E1 and P4502A6 in pigs. Meat Sci. 2006, 72, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Tambyrajah, W.; Doran, E.; Wood, J.; McGivan, J. The pig CYP2E1 promoter is activated by COUP-TF1 and HNF-1 and is inhibited by androstenone. Arch. Biochem. Biophys. 2004, 431, 252–260. [Google Scholar] [CrossRef]
- Liu, S.; Gonzalez, F. Role of the liver-enriched transcription factor HNF-1α in expression of the CYP2E1 gene. DNA Cell Biol. 2009, 14, 285–293. [Google Scholar] [CrossRef]
- Zadinová, K.; Stupka, R.; Stratil, A.; Čítek, J.; Vehovský, K.; Lebedová, N.; Šprysla, M.; Okrouhlá, M. Association analysis of SNPs in the porcine CYP2E1 gene with skatole, indole, and androstenone levels in backfat of a crossbred pig population. Meat Sci. 2017, 131, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Vogel, C.; Marcotte, E. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012, 13, 227–232. [Google Scholar] [CrossRef] [PubMed]
- De Sousa Abreu, R.; Penalva, L.; Marcotte, E.; Vogel, C. Global signatures of protein and mRNA expression levels. Mol. Biosyst. 2009, 5, 1512–1526. [Google Scholar] [CrossRef]
- Song, B.; Gelboin, H.; Park, S.; Yang, C.; Gonzalez, F. Complementary DNA and protein sequences of ethanol-inducible rat and human cytochrome P450s: Transcriptional and posttranscriptional regulation of the rat enzyme. J. Biol. Chem. 1986, 261, 16689–16697. [Google Scholar] [CrossRef] [PubMed]
- Puccinelli, E.; Gervasi, P.G.; Longo, V. Xenobiotic metabolizing cytochrome P450 in pig, a promising animal model. Curr. Drug Metab. 2011, 12, 507–525. [Google Scholar] [CrossRef]
- Brunius, C.; Ramussen, M.K.; Lacoutiere, H.; Andersson, K.; Ekstrand, B.; Zamaratskaia, G. Expression and activities of hepatic cytochrome P450 (CYP1A, CYP2A and CYP2E1) in entire and castrated male pigs. Animal 2012, 6, 271–277. [Google Scholar] [CrossRef]
| Breed | Carcass Weight (kg) | mRNA by qPCR (Fold Change) | Relative CYP2E1 Protein Expression |
|---|---|---|---|
| Duroc | 113.3 ± 4.35 a | 1.42 ± 1.19 a | 874.3 ± 68.7 a |
| Landrace | 112.4 ± 4.35 a | 1.30 ± 1.18 a | 956.3 ± 67.7 a |
| Yorkshire | 103.3 ± 3.89 a | 3.09 ± 1.03 a | 639.6 ± 59.4 b |
| SNP | Location from ATG Codon | Allele Counts | MAF | Mutation | ||
|---|---|---|---|---|---|---|
| 0 | 1 | 2 | ||||
| S35 | −159 | 49 | 12 | 4 | 0.154 | T > C |
| S36 | −586 | 20 | 35 | 10 | 0.423 | T > C |
| S102 | −1693 | 11 | 21 | 33 | 0.331 | C > T |
| S34 | −1806 | 10 | 35 | 20 | 0.423 | C > T |
| S81 | −2322 | 21 | 33 | 11 | 0.423 | A > T |
| S90 | −2369 | 37 | 23 | 5 | 0.254 | T > C |
| S103 | −2514 | 21 | 33 | 11 | 0.423 | A > G |
| SNP | Estimate 1 | Std. Error | Significance Level |
|---|---|---|---|
| S35 | −0.74 | 1.23 | 0.55 |
| S102 | −3.38 | 1.28 | 0.01 |
| S90 | −3.57 | 1.57 | 0.03 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Archer, H.; Soares, R.A.N.; Jafarikia, M.; Lillie, B.N.; Schenkel, F.; Squires, E.J. Effect of SNPs in the Promoter Region on the Expression of Cytochrome P450 2E1 (CYP2E1) in Pig Liver. Animals 2024, 14, 1163. https://doi.org/10.3390/ani14081163
Archer H, Soares RAN, Jafarikia M, Lillie BN, Schenkel F, Squires EJ. Effect of SNPs in the Promoter Region on the Expression of Cytochrome P450 2E1 (CYP2E1) in Pig Liver. Animals. 2024; 14(8):1163. https://doi.org/10.3390/ani14081163
Chicago/Turabian StyleArcher, Holly, Riani A. N. Soares, Mohsen Jafarikia, Brandon N. Lillie, Flavio Schenkel, and E. James Squires. 2024. "Effect of SNPs in the Promoter Region on the Expression of Cytochrome P450 2E1 (CYP2E1) in Pig Liver" Animals 14, no. 8: 1163. https://doi.org/10.3390/ani14081163
APA StyleArcher, H., Soares, R. A. N., Jafarikia, M., Lillie, B. N., Schenkel, F., & Squires, E. J. (2024). Effect of SNPs in the Promoter Region on the Expression of Cytochrome P450 2E1 (CYP2E1) in Pig Liver. Animals, 14(8), 1163. https://doi.org/10.3390/ani14081163






