Role of ADAR1 on Proliferation and Differentiation in Porcine Preadipocytes
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Cell Transfection
2.3. Flow Cytometry
2.4. Cell Counting Kit-8 Assay
2.5. EdU Staining
2.6. Oil Red O Staining
2.7. Triglyceride Determination
2.8. RT-qPCR Analysis
2.9. Western Blot Detection
2.10. RNA Sequencing and Data Processing
2.11. Statistical Analysis
3. Results
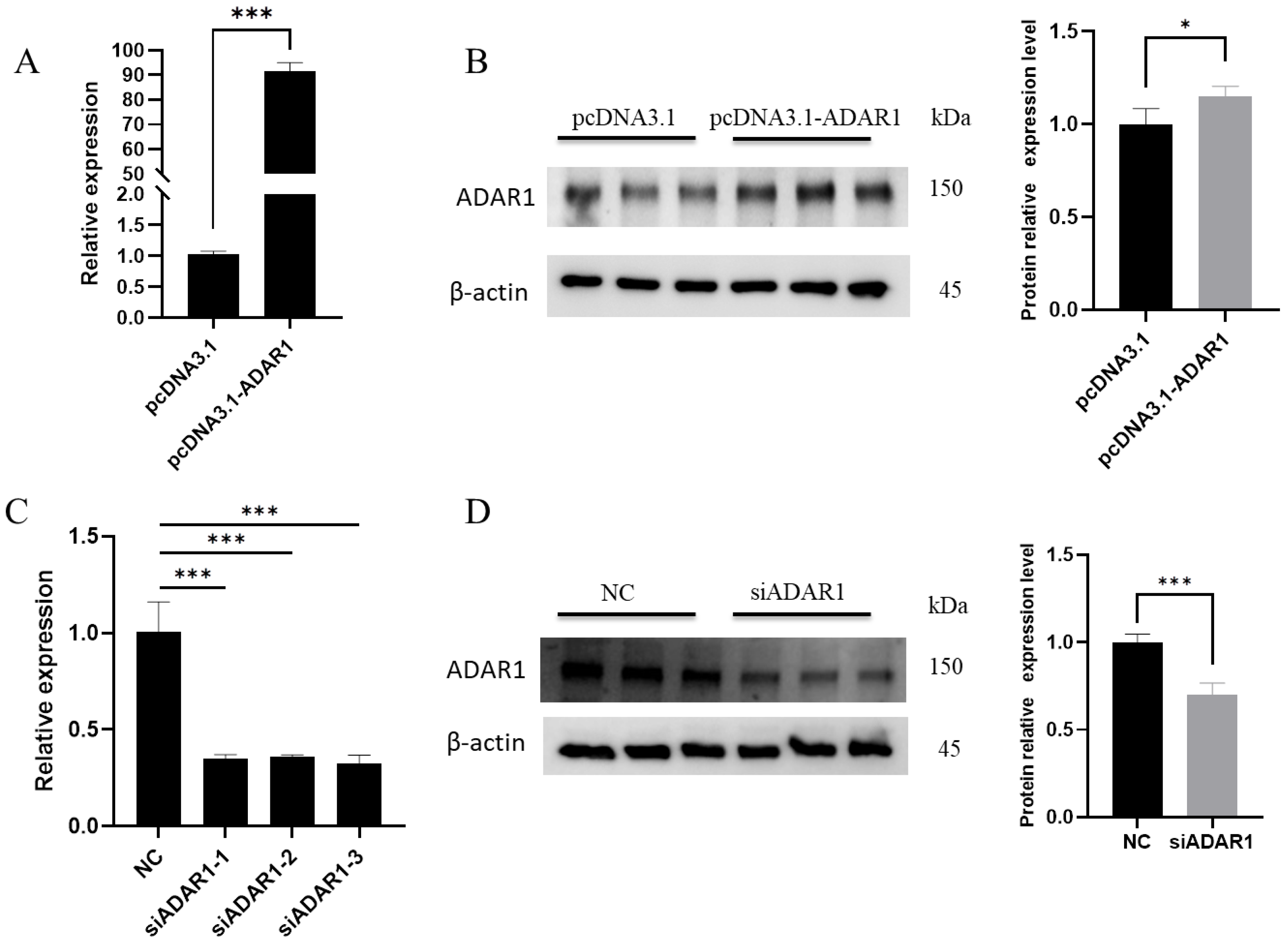
3.1. Efficiency of ADAR1 Interference and Overexpression
3.2. ADAR1 Promotes Proliferation of Porcine Preadipocytes
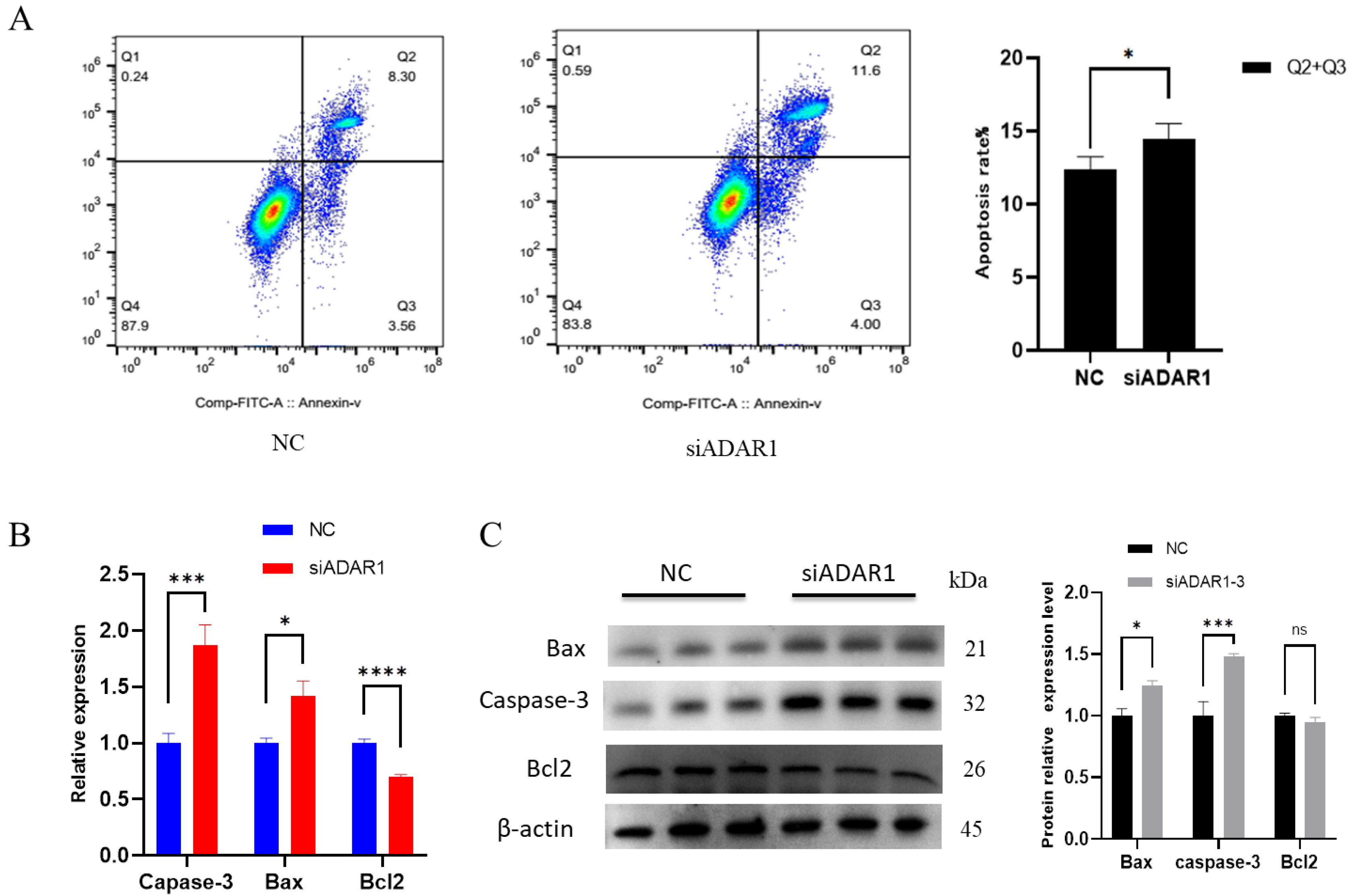
3.3. ADAR1 Inhibits Apoptosis of Porcine Preadipocytes
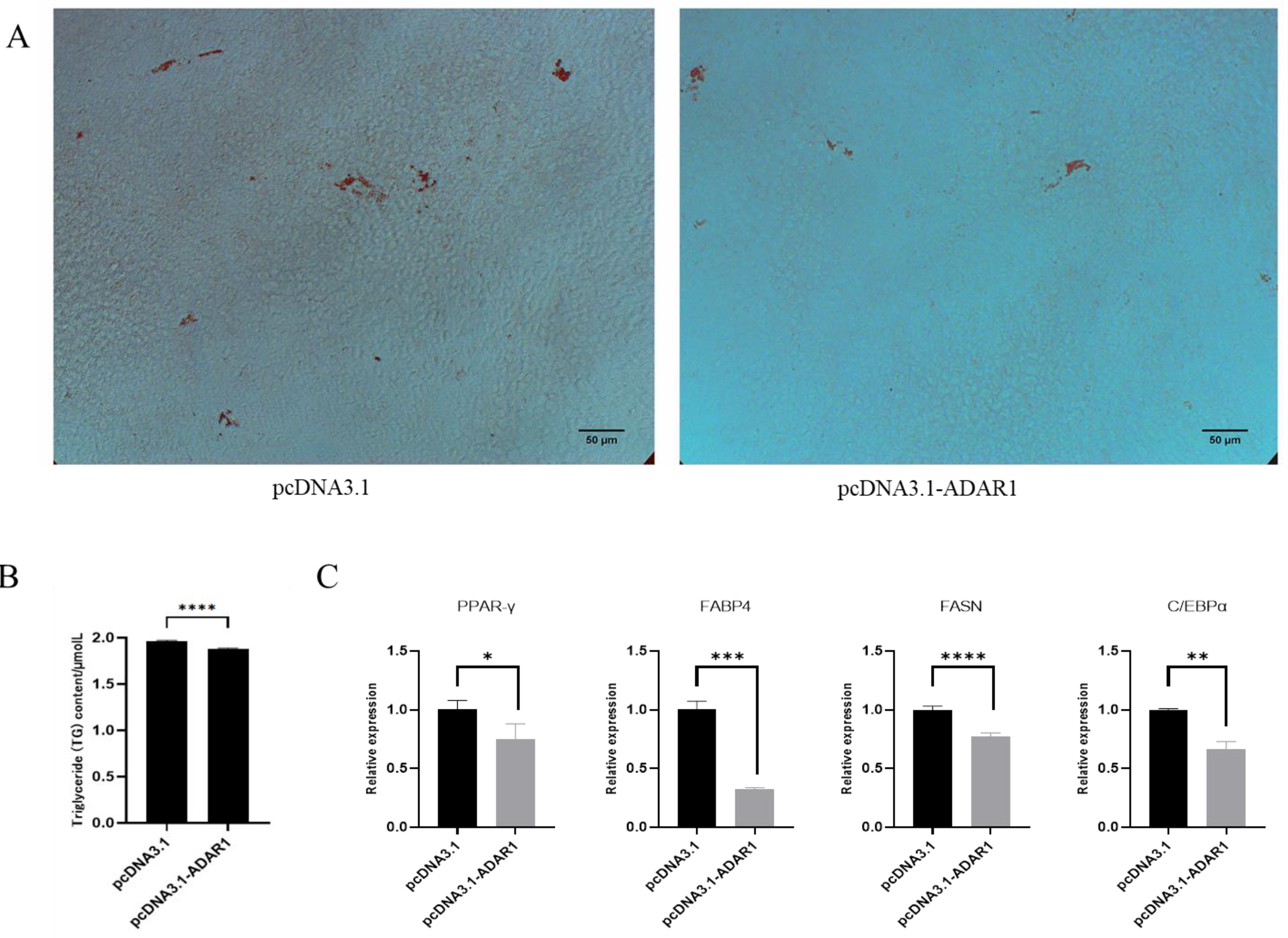
3.4. ADAR1 Inhibits Differentiation of Porcine Preadipocytes
3.5. Analysis of Differentially Expressed Genes (DEGs)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ghaben, A.L.; Scherer, P.E. Adipogenesis and metabolic health. Nat. Rev. Mol. Cell Biol. 2019, 20, 242–258. [Google Scholar] [CrossRef]
- Bass, B.L. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002, 71, 817–846. [Google Scholar] [CrossRef]
- Desterro, J.M.; Keegan, L.P.; Lafarga, M.; Berciano, M.T.; O’Connell, M.; Carmo-Fonseca, M. Dynamic association of RNA-editing enzymes with the nucleolus. J. Cell Sci. 2003, 116, 1805–1818. [Google Scholar] [CrossRef]
- Fei, J.; Cui, X.B.; Wang, J.N.; Dong, K.; Chen, S.Y. ADAR1-Mediated RNA Editing, A Novel Mechanism Controlling Phenotypic Modulation of Vascular Smooth Muscle Cells. Circ. Res. 2016, 119, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Guallar, D.; Fuentes-Iglesias, A.; Souto, Y.; Ameneiro, C.; Freire-Agulleiro, O.; Pardavila, J.A.; Escudero, A.; Garcia-Outeiral, V.; Moreira, T.; Saenz, C.; et al. ADAR1-Dependent RNA Editing Promotes MET and iPSC Reprogramming by Alleviating ER Stress. Cell Stem Cell 2020, 27, 300–314. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Liu, S.; Yan, R.; Nguyen, V.; Zenati, M.; Billiar, T.R.; Wang, Q. ADAR1 RNA editing regulates endothelial cell functions via the MDA-5 RNA sensing signaling pathway. Life Sci. Alliance 2022, 5, e202101191. [Google Scholar] [CrossRef] [PubMed]
- Lamers, M.M.; van den Hoogen, B.G.; Haagmans, B.L. ADAR1: “Editor-in-Chief” of Cytoplasmic Innate Immunity. Front. Immunol. 2019, 10, 1763. [Google Scholar] [CrossRef]
- Widmark, A.; Sagredo, E.A.; Karlstrom, V.; Behm, M.; Biryukova, I.; Friedlander, M.R.; Daniel, C.; Ohman, M. ADAR1- and ADAR2-mediated regulation of maturation and targeting of miR-376b to modulate GABA neurotransmitter catabolism. J. Biol. Chem. 2022, 298, 101682. [Google Scholar] [CrossRef]
- Shen, P.; Yang, T.; Chen, Q.; Yuan, H.; Wu, P.; Cai, B.; Meng, L.; Huang, X.; Liu, J.; Zhang, Y.; et al. CircNEIL3 regulatory loop promotes pancreatic ductal adenocarcinoma progression via miRNA sponging and A-to-I RNA-editing. Mol. Cancer 2021, 20, 51. [Google Scholar] [CrossRef]
- Ramirez-Moya, J.; Miliotis, C.; Baker, A.R.; Gregory, R.I.; Slack, F.J.; Santisteban, P. An ADAR1-dependent RNA editing event in the cyclin-dependent kinase CDK13 promotes thyroid cancer hallmarks. Mol. Cancer 2021, 20, 115. [Google Scholar] [CrossRef]
- Chen, L.; Li, Y.; Lin, C.H.; Chan, T.H.; Chow, R.K.; Song, Y.; Liu, M.; Yuan, Y.F.; Fu, L.; Kong, K.L.; et al. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat. Med. 2013, 19, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Luo, R.; Li, Y.; Li, X.; Yang, Z.; Peng, J.; Huang, K. ADAR1 inhibits adipogenesis and obesity by interacting with Dicer to promote the maturation of miR-155-5P. J. Cell Sci. 2022, 135, 259333. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, L.; Yue, J.; Wei, X.; Wang, L.; Liu, X.; Gao, H.; Hou, X.; Zhao, F.; Yan, H.; et al. Genome-wide identification of RNA editing in seven porcine tissues by matched DNA and RNA high-throughput sequencing. J. Anim. Sci. Biotechnol. 2019, 10, 24. [Google Scholar] [CrossRef] [PubMed]
- Baker, A.R.; Slack, F.J. ADAR1 and its implications in cancer development and treatment. Trends Genet. 2022, 38, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, Y.; Zhang, J.; Liu, L.; Chen, Y.; Yang, X.; Liao, X.; He, M.; Jia, Z.; Fan, J.; et al. ADAR1 regulates vascular remodeling in hypoxic pulmonary hypertension through N1-methyladenosine modification of circCDK17. Acta Pharm. Sin. B 2023, 13, 4840–4855. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Hu, P.; Lin, X.; Han, W.; Zhu, L.; Tan, X.; Ye, F.; Wang, G.; Wu, F.; Yin, B.; et al. PTBP1 induces ADAR1 p110 isoform expression through IRES-like dependent translation control and influences cell proliferation in gliomas. Cell Mol. Life Sci. 2015, 72, 4383–4397. [Google Scholar] [CrossRef]
- Xiao, H.; Cheng, Q.; Wu, X.; Tang, Y.; Liu, J.; Li, X. ADAR1 may be involved in the proliferation of acute myeloid leukemia cells via regulation of the Wnt pathway. Cancer Manag. Res. 2019, 11, 8547–8555. [Google Scholar] [CrossRef]
- Bybee, A.; Thomas, N.S. Cell cycle regulation. Blood Rev. 1991, 5, 177–192. [Google Scholar] [CrossRef]
- Zhang, F.; Rabinovici, R. Adenosine deaminase acting on RNA 1 accelerates cell cycle through increased translation and activity of cyclin-dependent kinase 2. Shock 2007, 27, 214–219. [Google Scholar] [CrossRef]
- Maga, G.; Hubscher, U. Proliferating cell nuclear antigen (PCNA): A dancer with many partners. J. Cell Sci. 2003, 116, 3051–3060. [Google Scholar] [CrossRef]
- Bravo, R.; Frank, R.; Blundell, P.A.; Macdonald-Bravo, H. Cyclin/PCNA is the auxiliary protein of DNA polymerase-delta. Nature 1987, 326, 515–517. [Google Scholar] [CrossRef] [PubMed]
- UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [CrossRef] [PubMed]
- Li, X.; Zhu, R.; Yuan, Y.; Cai, Z.; Liang, S.; Bian, J.; Xu, G. Double-stranded RNA-specific adenosine deaminase-knockdown inhibits the proliferation and induces apoptosis of DU145 and PC3 cells by promoting the phosphorylation of H2A.X variant histone. Oncol. Lett. 2021, 22, 764. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Wang, Z.; Chen, X.; Wang, W.; Wang, X. ADAR1 silencing-induced HUVEC apoptosis is mediated by FGFR2 under hypoxia stress. Drug Des. Devel. Ther. 2018, 12, 4181–4189. [Google Scholar] [CrossRef] [PubMed]
- Katayama, S.; Shimoda, K.; Takenaga, Y. Loss of ADAR1 in human iPS cells promotes caspase3-mediated apoptotic cell death. Genes. Cells 2015, 20, 675–680. [Google Scholar] [CrossRef] [PubMed]
- Barneda, D.; Christian, M. Lipid droplet growth: Regulation of a dynamic organelle. Curr. Opin. Cell Biol. 2017, 47, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Semenkovich, C.F. Regulation of fatty acid synthase (FAS). Prog. Lipid Res. 1997, 36, 43–53. [Google Scholar] [CrossRef]
- Ntambi, J.M.; Young-Cheul, K. Adipocyte differentiation and gene expression. J. Nutr. 2000, 130, 3122S–3126S. [Google Scholar] [CrossRef]
- MacDougald, O.A.; Lane, M.D. Transcriptional regulation of gene expression during adipocyte differentiation. Annu. Rev. Biochem. 1995, 64, 345–373. [Google Scholar] [CrossRef]
- Chung, Y.C.; Hyun, C.G. Inhibitory Effects of Pinostilbene on Adipogenesis in 3T3-L1 Adipocytes: A Study of Possible Mechanisms. Int. J. Mol. Sci. 2021, 22, 13446. [Google Scholar] [CrossRef]
- Chen, S.; Li, Z.; Li, W.; Shan, Z.; Zhu, W. Resveratrol inhibits cell differentiation in 3T3-L1 adipocytes via activation of AMPK. Can. J. Physiol. Pharm. 2011, 89, 793–799. [Google Scholar] [CrossRef]
- Ha, D.T.; Trung, T.N.; Phuong, T.T.; Yim, N.; Chen, Q.C.; Bae, K. The selected flavonol glycoside derived from Sophorae Flos improves glucose uptake and inhibits adipocyte differentiation via activation AMPK in 3T3-L1 cells. Bioorganic Med. Chem. Lett. 2010, 20, 6076–6081. [Google Scholar] [CrossRef]
- Zuniga, L.A.; Shen, W.J.; Joyce-Shaikh, B.; Pyatnova, E.A.; Richards, A.G.; Thom, C.; Andrade, S.M.; Cua, D.J.; Kraemer, F.B.; Butcher, E.C. IL-17 regulates adipogenesis, glucose homeostasis, and obesity. J. Immunol. 2010, 185, 6947–6959. [Google Scholar] [CrossRef] [PubMed]
- Hardwick, J.P.; Eckman, K.; Lee, Y.K.; Abdelmegeed, M.A.; Esterle, A.; Chilian, W.M.; Chiang, J.Y.; Song, B.J. Eicosanoids in metabolic syndrome. Adv. Pharmacol. 2013, 66, 157–266. [Google Scholar] [CrossRef] [PubMed]
- Pulcinelli, F.M.; Biasucci, L.M.; Riondino, S.; Giubilato, S.; Leo, A.; Di Renzo, L.; Trifiro, E.; Mattiello, T.; Pitocco, D.; Liuzzo, G.; et al. COX-1 sensitivity and thromboxane A2 production in type 1 and type 2 diabetic patients under chronic aspirin treatment. Eur. Heart J. 2009, 30, 1279–1286. [Google Scholar] [CrossRef]
- Graziani, F.; Biasucci, L.M.; Cialdella, P.; Liuzzo, G.; Giubilato, S.; Della, B.R.; Pulcinelli, F.M.; Iaconelli, A.; Mingrone, G.; Crea, F. Thromboxane production in morbidly obese subjects. Am. J. Cardiol. 2011, 107, 1656–1661. [Google Scholar] [CrossRef]
- Song, S.H.; Kim, K.; Jo, E.K.; Kim, Y.W.; Kwon, J.S.; Bae, S.S.; Sung, J.H.; Park, S.G.; Kim, J.T.; Suh, W. Fibroblast Growth Factor 12 Is a Novel Regulator of Vascular Smooth Muscle Cell Plasticity and Fate. Arterioscl Throm Vas. 2016, 36, 1928–1936. [Google Scholar] [CrossRef]
- Yan, Q.; Fang, X.; Liu, X.; Guo, S.; Chen, S.; Luo, M.; Lan, P.; Guan, X.Y. Loss of ESRP2 Activates TAK1-MAPK Signaling through the Fetal RNA-Splicing Program to Promote Hepatocellular Carcinoma Progression. Adv. Sci. 2024, 11, e2305653. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, M.; Jiang, J.; Ren, R.; Gao, N.; He, J.; Zhang, Y. Role of ADAR1 on Proliferation and Differentiation in Porcine Preadipocytes. Animals 2024, 14, 1201. https://doi.org/10.3390/ani14081201
Yang M, Jiang J, Ren R, Gao N, He J, Zhang Y. Role of ADAR1 on Proliferation and Differentiation in Porcine Preadipocytes. Animals. 2024; 14(8):1201. https://doi.org/10.3390/ani14081201
Chicago/Turabian StyleYang, Menghuan, Jun Jiang, Ruimin Ren, Ning Gao, Jun He, and Yuebo Zhang. 2024. "Role of ADAR1 on Proliferation and Differentiation in Porcine Preadipocytes" Animals 14, no. 8: 1201. https://doi.org/10.3390/ani14081201




