A Strategy for Single-Run Sequencing of the Water Buffalo Genome: (II) Fast One-Step Assembly of Highly Continuous Chromosome Sequences
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sequencing by Long Reads Strategy
2.2. Read Preprocessing and Genome Assembly
2.3. Assembly Statistics and Reference-Based Evaluation
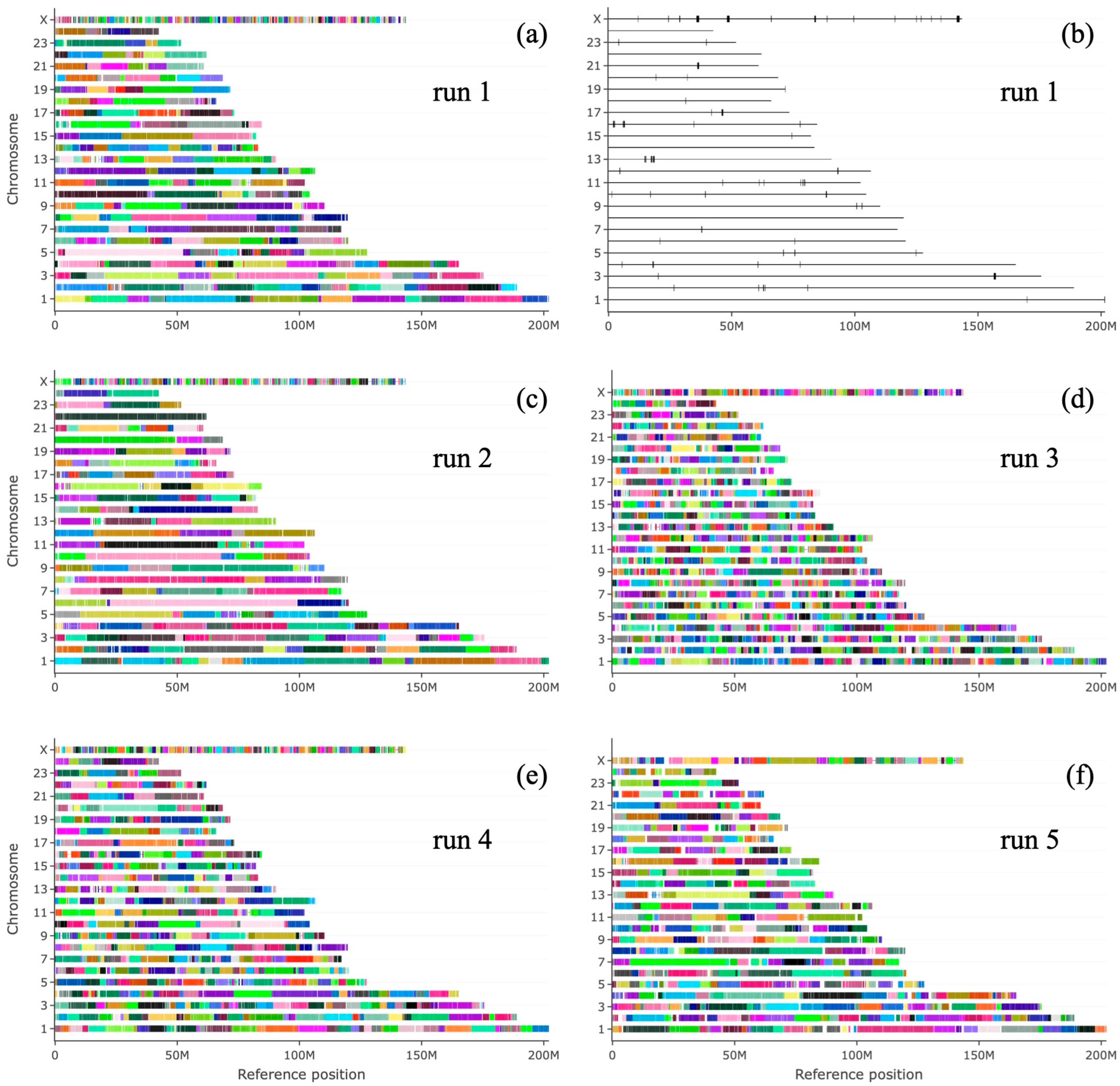
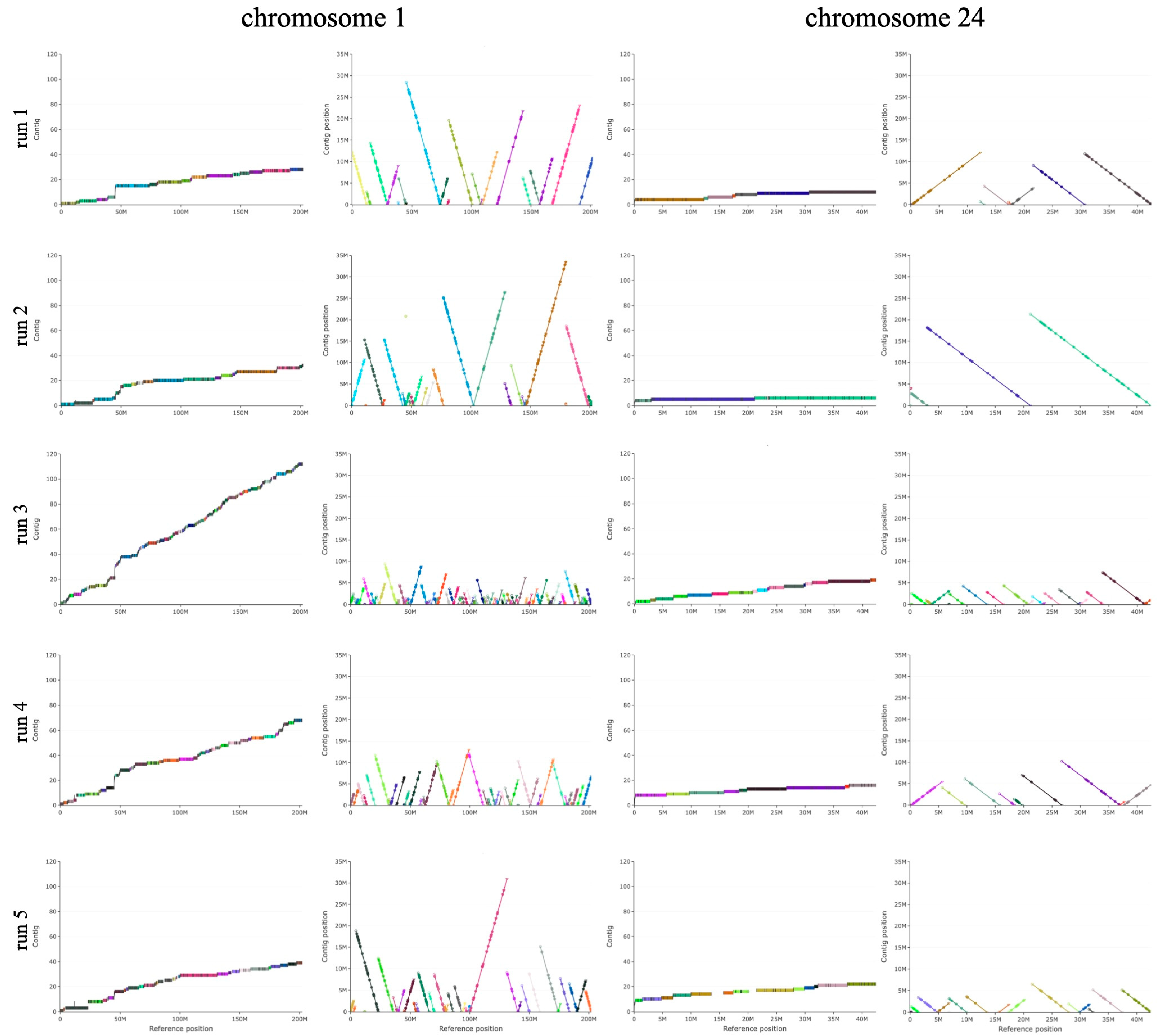
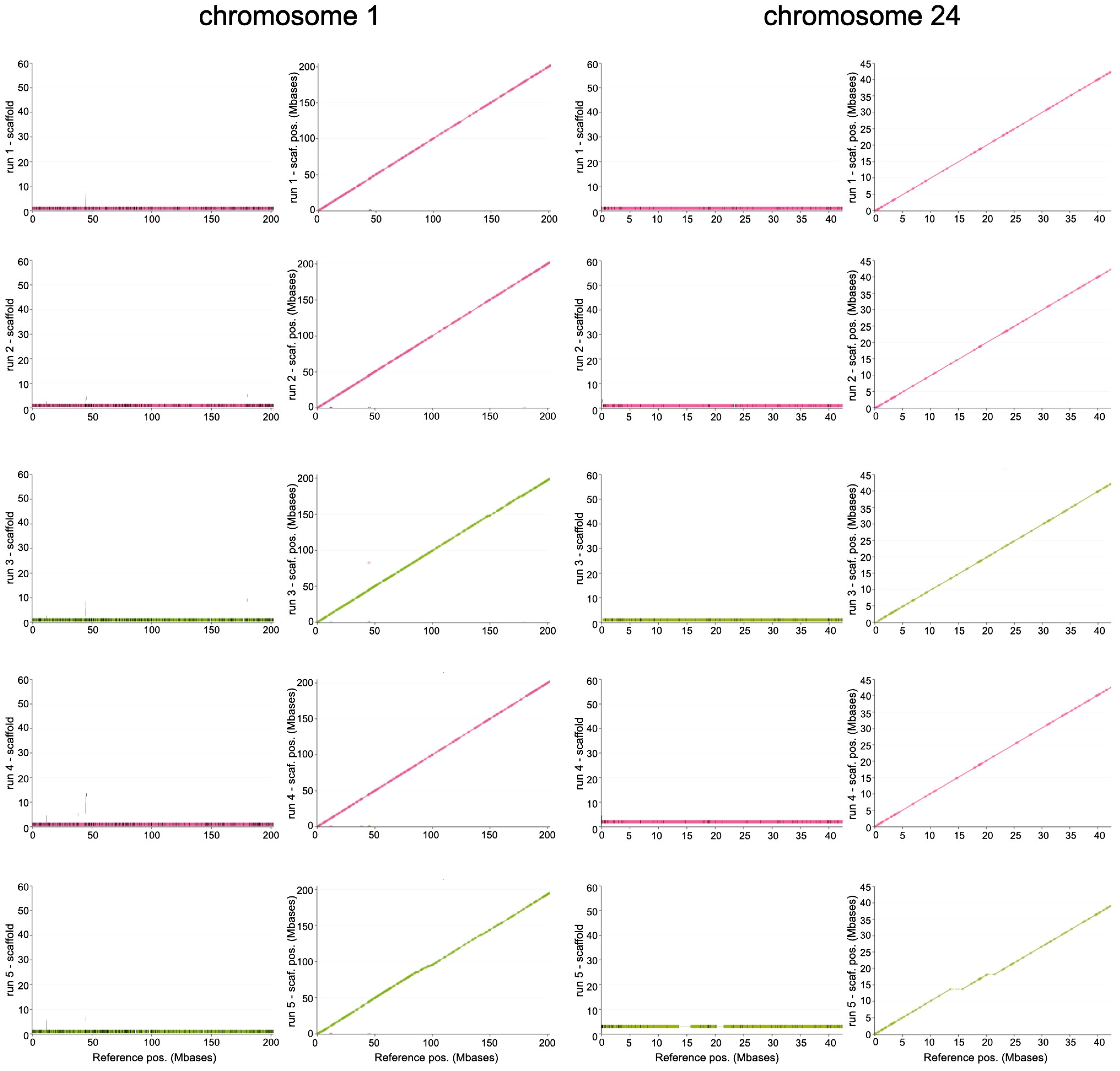
3. Results
3.1. De Novo Genome Assembly from a Single Run of Long-Read Sequencing
3.2. Reference-Driven Scaffolding Step Results in Good-Quality Genome
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| SNP | Single Nucleotide Polymorphism |
| WGS | Whole-Genome Sequencing |
| SV | Structural Variation |
| CNV | Copy Number Variation |
References
- Gutierrez-Reinoso, M.A.; Aponte, P.M.; Garcia-Herreros, M. Genomic Analysis, Progress and Future Perspectives in Dairy Cattle Selection: A Review. Animals 2021, 11, 599. [Google Scholar] [CrossRef]
- Jones, H.E.; Wilson, P.B. Progress and Opportunities through Use of Genomics in Animal Production. Trends Genet. 2022, 38, 1228–1252. [Google Scholar] [CrossRef]
- Wiggans, G.R.; Cole, J.B.; Hubbard, S.M.; Sonstegard, T.S. Genomic Selection in Dairy Cattle: The USDA Experience. Annu. Rev. Anim. Biosci. 2017, 5, 309–327. [Google Scholar] [CrossRef] [PubMed]
- Elsik, C.G.; Tellam, R.L.; Worley, K.C. The Genome Sequence of Taurine Cattle: A Window to Ruminant Biology and Evolution. Science 2009, 324, 522–528. [Google Scholar] [CrossRef] [PubMed]
- Kadarmideen, H.N. Genomics to Systems Biology in Animal and Veterinary Sciences: Progress, Lessons and Opportunities. Livest. Sci. 2014, 166, 232–248. [Google Scholar] [CrossRef]
- Guarini, A.R.; Lourenco, D.A.L.; Brito, L.F.; Sargolzaei, M.; Baes, C.F.; Miglior, F.; Tsuruta, S.; Misztal, I.; Schenkel, F.S. Use of a Single-Step Approach for Integrating Foreign Information into National Genomic Evaluation in Holstein Cattle. J. Dairy. Sci. 2019, 102, 8175–8183. [Google Scholar] [CrossRef]
- Lombardo, B.; Pagani, M.; De Rosa, A.; Nunziato, M.; Migliarini, S.; Garofalo, M.; Terrile, M.; D’Argenio, V.; Galbusera, A.; Nuzzo, T.; et al. D-Aspartate Oxidase Gene Duplication Induces Social Recognition Memory Deficit in Mice and Intellectual Disabilities in Humans. Transl. Psychiatry 2022, 12, 305. [Google Scholar] [CrossRef] [PubMed]
- Hayes, B.J.; Daetwyler, H.D. 1000 Bull Genomes Project to Map Simple and Complex Genetic Traits in Cattle: Applications and Outcomes. Annu. Rev. Anim. Biosci. 2019, 7, 89–102. [Google Scholar] [CrossRef]
- Della Coletta, R.; Qiu, Y.; Ou, S.; Hufford, M.B.; Hirsch, C.N. How the Pan-Genome Is Changing Crop Genomics and Improvement. Genome Biol. 2021, 22, 3. [Google Scholar] [CrossRef]
- Gong, Y.; Li, Y.; Liu, X.; Ma, Y.; Jiang, L. A Review of the Pangenome: How It Affects Our Understanding of Genomic Variation, Selection and Breeding in Domestic Animals? J. Anim. Sci. Biotechnol. 2023, 14, 73. [Google Scholar] [CrossRef]
- Zhu, F.; Yin, Z.-T.; Wang, Z.; Smith, J.; Zhang, F.; Martin, F.; Ogeh, D.; Hincke, M.; Lin, F.-B.; Burt, D.W.; et al. Three Chromosome-Level Duck Genome Assemblies Provide Insights into Genomic Variation during Domestication. Nat. Commun. 2021, 12, 5932. [Google Scholar] [CrossRef]
- Huang, Z.; Xu, Z.; Bai, H.; Huang, Y.; Kang, N.; Ding, X.; Liu, J.; Luo, H.; Yang, C.; Chen, W.; et al. Evolutionary Analysis of a Complete Chicken Genome. Proc. Natl. Acad. Sci. USA 2023, 120, e2216641120. [Google Scholar] [CrossRef]
- Zhu, F.; Yin, Z.-T.; Zhao, Q.-S.; Sun, Y.-X.; Jie, Y.-C.; Smith, J.; Yang, Y.-Z.; Burt, D.W.; Hincke, M.; Zhang, Z.-D.; et al. A Chromosome-Level Genome Assembly for the Silkie Chicken Resolves Complete Sequences for Key Chicken Metabolic, Reproductive, and Immunity Genes. Commun. Biol. 2023, 6, 1233. [Google Scholar] [CrossRef]
- Leonard, A.S.; Crysnanto, D.; Fang, Z.-H.; Heaton, M.P.; Vander Ley, B.L.; Herrera, C.; Bollwein, H.; Bickhart, D.M.; Kuhn, K.L.; Smith, T.P.L.; et al. Structural Variant-Based Pangenome Construction Has Low Sensitivity to Variability of Haplotype-Resolved Bovine Assemblies. Nat. Commun. 2022, 13, 3012. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Bian, P.; Hu, D.; Luo, F.; Huang, Y.; Jiao, S.; Wang, X.; Gong, M.; Li, R.; Cai, Y.; et al. A Chinese Indicine Pangenome Reveals a Wealth of Novel Structural Variants Introgressed from Other Bos Species. Genome Res. 2023, 33, 1284–1298. [Google Scholar] [CrossRef] [PubMed]
- Low, W.Y.; Tearle, R.; Bickhart, D.M.; Rosen, B.D.; Kingan, S.B.; Swale, T.; Thibaud-Nissen, F.; Murphy, T.D.; Young, R.; Lefevre, L.; et al. Chromosome-Level Assembly of the Water Buffalo Genome Surpasses Human and Goat Genomes in Sequence Contiguity. Nat. Commun. 2019, 10, 260. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Zhou, Y.; Zhang, B.; Zhang, Y.; Wang, X.; Feng, T.; Li, Z.; Cui, K.; Wang, Z.; Luo, C.; et al. Understanding Divergent Domestication Traits from the Whole-Genome Sequencing of Swamp- and River-Buffalo Populations. Natl. Sci. Rev. 2020, 7, 686–701. [Google Scholar] [CrossRef]
- Williams, J.L.; Iamartino, D.; Pruitt, K.D.; Sonstegard, T.; Smith, T.P.L.; Low, W.Y.; Biagini, T.; Bomba, L.; Capomaccio, S.; Castiglioni, B.; et al. Genome Assembly and Transcriptome Resource for River Buffalo, Bubalus bubalis (2n = 50). GigaScience 2017, 6, gix088. [Google Scholar] [CrossRef]
- Liu, X.; Liu, W.; Lenstra, J.A.; Zheng, Z.; Wu, X.; Yang, J.; Li, B.; Yang, Y.; Qiu, Q.; Liu, H.; et al. Evolutionary Origin of Genomic Structural Variations in Domestic Yaks. Nat. Commun. 2023, 14, 5617. [Google Scholar] [CrossRef]
- Gao, X.; Wang, S.; Wang, Y.-F.; Li, S.; Wu, S.-X.; Yan, R.-G.; Zhang, Y.-W.; Wan, R.-D.; He, Z.; Song, R.-D.; et al. Long Read Genome Assemblies Complemented by Single Cell RNA-Sequencing Reveal Genetic and Cellular Mechanisms Underlying the Adaptive Evolution of Yak. Nat. Commun. 2022, 13, 4887. [Google Scholar] [CrossRef]
- Jiang, Y.-F.; Wang, S.; Wang, C.-L.; Xu, R.-H.; Wang, W.-W.; Jiang, Y.; Wang, M.-S.; Jiang, L.; Dai, L.-H.; Wang, J.-R.; et al. Pangenome Obtained by Long-Read Sequencing of 11 Genomes Reveal Hidden Functional Structural Variants in Pigs. iScience 2023, 26, 106119. [Google Scholar] [CrossRef]
- Warr, A.; Affara, N.; Aken, B.; Beiki, H.; Bickhart, D.M.; Billis, K.; Chow, W.; Eory, L.; Finlayson, H.A.; Flicek, P.; et al. An Improved Pig Reference Genome Sequence to Enable Pig Genetics and Genomics Research. Gigascience 2020, 9, giaa051. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Yang, P.; Dai, X.; Asadollahpour Nanaei, H.; Fang, W.; Yang, Z.; Cai, Y.; Zheng, Z.; Wang, X.; Jiang, Y. A near Complete Genome for Goat Genetic and Genomic Research. Genet. Sel. Evol. 2021, 53, 74. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wu, Y.; Chen, B.; Cai, Y.; Guo, J.; Leonard, A.S.; Kalds, P.; Zhou, S.; Zhang, J.; Zhou, P.; et al. Markhor-Derived Introgression of a Genomic Region Encompassing PAPSS2 Confers High-Altitude Adaptability in Tibetan Goats. Mol. Biol. Evol. 2022, 39, msac253. [Google Scholar] [CrossRef] [PubMed]
- Qiao, G.; Xu, P.; Guo, T.; Wu, Y.; Lu, X.; Zhang, Q.; He, X.; Zhu, S.; Zhao, H.; Lei, Z.; et al. Genetic Basis of Dorper Sheep (Ovis Aries) Revealed by Long-Read De Novo Genome Assembly. Front. Genet. 2022, 13, 846449. [Google Scholar] [CrossRef]
- Li, R.; Gong, M.; Zhang, X.; Wang, F.; Liu, Z.; Zhang, L.; Yang, Q.; Xu, Y.; Xu, M.; Zhang, H.; et al. A Sheep Pangenome Reveals the Spectrum of Structural Variations and Their Effects on Tail Phenotypes. Genome Res. 2023, 33, 463–477. [Google Scholar] [CrossRef]
- Liu, X.; Zheng, J.; Ding, J.; Wu, J.; Zuo, F.; Zhang, G. When Livestock Genomes Meet Third-Generation Sequencing Technology: From Opportunities to Applications. Genes 2024, 15, 245. [Google Scholar] [CrossRef]
- Satam, H.; Joshi, K.; Mangrolia, U.; Waghoo, S.; Zaidi, G.; Rawool, S.; Thakare, R.P.; Banday, S.; Mishra, A.K.; Das, G.; et al. Next-Generation Sequencing Technology: Current Trends and Advancements. Biology 2023, 12, 997. [Google Scholar] [CrossRef]
- Goldfeder, R.L.; Wall, D.P.; Khoury, M.J.; Ioannidis, J.P.A.; Ashley, E.A. Human Genome Sequencing at the Population Scale: A Primer on High-Throughput DNA Sequencing and Analysis. Am. J. Epidemiol. 2017, 186, 1000–1009. [Google Scholar] [CrossRef]
- Di Maggio, F.; Nunziato, M.; Toscano, E.; Sepe, L.; Cimmino, R.; Capolongo, E.A.; Vasco, A.; Paolella, G.; Salvatore, F. A Strategy for Single-Run Sequencing of the Water Buffalo Genome: (I) the Use of Third-Generation Technology to Quickly Produce Long, High-Quality Reads. Animals 2025, 15, 2991. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Completing Bacterial Genome Assemblies with Multiplex MinION Sequencing. Microb. Genom. 2017, 3, e000132. [Google Scholar] [CrossRef]
- Shafin, K.; Pesout, T.; Lorig-Roach, R.; Haukness, M.; Olsen, H.E.; Bosworth, C.; Armstrong, J.; Tigyi, K.; Maurer, N.; Koren, S.; et al. Nanopore Sequencing and the Shasta Toolkit Enable Efficient de Novo Assembly of Eleven Human Genomes. Nat. Biotechnol. 2020, 38, 1044–1053. [Google Scholar] [CrossRef]
- Alonge, M.; Lebeigle, L.; Kirsche, M.; Jenike, K.; Ou, S.; Aganezov, S.; Wang, X.; Lippman, Z.B.; Schatz, M.C.; Soyk, S. Automated Assembly Scaffolding Using RagTag Elevates a New Tomato System for High-Throughput Genome Editing. Genome Biol. 2022, 23, 258. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality Assessment Tool for Genome Assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Mikheenko, A.; Prjibelski, A.; Saveliev, V.; Antipov, D.; Gurevich, A. Versatile Genome Assembly Evaluation with QUAST-LG. Bioinformatics 2018, 34, i142–i150. [Google Scholar] [CrossRef] [PubMed]
- Toscano, E.; Sepe, L.; Del Giudice, G.; Tufano, R.; Paolella, G. A Three Component Model for Superdiffusive Motion Effectively Describes Migration of Eukaryotic Cells Moving Freely or under a Directional Stimulus. PLoS ONE 2022, 17, e0272259. [Google Scholar] [CrossRef] [PubMed]
- Catapano, R.; Sepe, L.; Toscano, E.; Paolella, G.; Chiurazzi, F.; Barbato, S.P.; Bruzzese, D.; Arianna, R.; Grosso, M.; Romano, S.; et al. Biological Relevance of ZNF224 Expression in Chronic Lymphocytic Leukemia and Its Implication IN NF-kB Pathway Regulation. Front. Mol. Biosci. 2022, 9, 1010984. [Google Scholar] [CrossRef]
- Sepe, L.; Candia, U.; Sasso del Verme, D.; Toscano, E.; Toriello, M.; Sodaro, G.; Rapuano, R.; Romano, S.; Grosso, M.; Paolella, G.; et al. ZNF224 Enhances the Oncogenic Function of P21 via P53 and AKT Pathways in Melanoma. FEBS J. 2025, 292, 3986–4005. [Google Scholar] [CrossRef] [PubMed]
- Jing, X.; Emerson, M.; Masters, D.; Brooks, M.; Buskirk, J.; Abukamail, N.; Liu, C.; Cimino, J.J.; Shubrook, J.; De Lacalle, S.; et al. A Visual Interactive Analytic Tool for Filtering and Summarizing Large Health Data Sets Coded with Hierarchical Terminologies (VIADS). BMC Med. Inf. Inform. Decis. Mak. 2019, 19, 31. [Google Scholar] [CrossRef]
- Toscano, E.; Cimmino, E.; Boccia, A.; Sepe, L.; Paolella, G. Cell Populations Simulated in Silico within SimulCell Accurately Reproduce the Behaviour of Experimental Cell Cultures. npj Syst. Biol. Appl. 2025, 11, 48. [Google Scholar] [CrossRef]
- Iannuzzi, L.; Di Meo, G.P.; Perucatti, A.; Ferrara, L. The High Resolution G- and R-Banding Pattern in Chromosomes of River Buffalo (Bubalus bubalis L.). Hereditas 1990, 112, 209–215. [Google Scholar] [CrossRef]
- El Nahas, S.M.; de Hondt, H.A.; Womack, J.E. Current Status of the River Buffalo (Bubalus bubalis L.) Gene Map. J. Hered. 2001, 92, 221–225. [Google Scholar] [CrossRef]
- Logsdon, G.A.; Rozanski, A.N.; Ryabov, F.; Potapova, T.; Shepelev, V.A.; Catacchio, C.R.; Porubsky, D.; Mao, Y.; Yoo, D.; Rautiainen, M.; et al. The Variation and Evolution of Complete Human Centromeres. Nature 2024, 629, 136–145. [Google Scholar] [CrossRef]
- Kubickova, S.; Kopecna, O.; Cernohorska, H.; Rubes, J.; Vozdova, M. X Chromosome-Specific Repeats in Non-Domestic Bovidae. Genes 2024, 15, 159. [Google Scholar] [CrossRef] [PubMed]
- Miga, K.H.; Koren, S.; Rhie, A.; Vollger, M.R.; Gershman, A.; Bzikadze, A.; Brooks, S.; Howe, E.; Porubsky, D.; Logsdon, G.A.; et al. Telomere-to-Telomere Assembly of a Complete Human X Chromosome. Nature 2020, 585, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Genome Assembly in the Telomere-to-Telomere Era. Nat. Rev. Genet. 2024, 25, 658–670. [Google Scholar] [CrossRef]
- Egger-Danner, C.; Cole, J.B.; Pryce, J.E.; Gengler, N.; Heringstad, B.; Bradley, A.; Stock, K.F. Invited Review: Overview of New Traits and Phenotyping Strategies in Dairy Cattle with a Focus on Functional Traits. Animal 2015, 9, 191–207. [Google Scholar] [CrossRef] [PubMed]
- Miglior, F.; Fleming, A.; Malchiodi, F.; Brito, L.F.; Martin, P.; Baes, C.F. A 100-Year Review: Identification and Genetic Selection of Economically Important Traits in Dairy Cattle. J. Dairy. Sci. 2017, 100, 10251–10271. [Google Scholar] [CrossRef]
- Sun, H.Z.; Plastow, G.; Guan, L.L. Invited Review: Advances and Challenges in Application of Feedomics to Improve Dairy Cow Production and Health. J. Dairy. Sci. 2019, 102, 5853–5870. [Google Scholar] [CrossRef]
- García-Ruiz, A.; Cole, J.B.; VanRaden, P.M.; Wiggans, G.R.; Ruiz-López, F.J.; Van Tassell, C.P. Changes in Genetic Selection Differentials and Generation Intervals in US Holstein Dairy Cattle as a Result of Genomic Selection. Proc. Natl. Acad. Sci. USA 2016, 113, E3995–E4004. [Google Scholar] [CrossRef]
- Berry, D.P.; Wall, E.; Pryce, J.E. Genetics and Genomics of Reproductive Performance in Dairy and Beef Cattle. Animal 2014, 8 (Suppl. 1), 105–121. [Google Scholar] [CrossRef] [PubMed]
- Giuffrida, G.; D’Argenio, V.; Ferraù, F.; Lasorsa, V.A.; Polito, F.; Aliquò, F.; Ragonese, M.; Cotta, O.R.; Alessi, Y.; Oteri, R.; et al. Methylome Analysis in Nonfunctioning and GH-Secreting Pituitary Adenomas. Front. Endocrinol. 2022, 13, 841118. [Google Scholar] [CrossRef] [PubMed]
- Peona, V.; Blom, M.P.K.; Xu, L.; Burri, R.; Sullivan, S.; Bunikis, I.; Liachko, I.; Haryoko, T.; Jønsson, K.A.; Zhou, Q.; et al. Identifying the Causes and Consequences of Assembly Gaps Using a Multiplatform Genome Assembly of a Bird-of-Paradise. Mol. Ecol. Resour. 2021, 21, 263–286. [Google Scholar] [CrossRef]
- Nurk, S.; Koren, S.; Rhie, A.; Rautiainen, M.; Bzikadze, A.V.; Mikheenko, A.; Vollger, M.R.; Altemose, N.; Uralsky, L.; Gershman, A.; et al. The Complete Sequence of a Human Genome. Science 2022, 376, 44–53. [Google Scholar] [CrossRef]
- Larsen, P.A.; Harris, R.A.; Liu, Y.; Murali, S.C.; Campbell, C.R.; Brown, A.D.; Sullivan, B.A.; Shelton, J.; Brown, S.J.; Raveendran, M.; et al. Hybrid de Novo Genome Assembly and Centromere Characterization of the Gray Mouse Lemur (Microcebus murinus). BMC Biol. 2017, 15, 110. [Google Scholar] [CrossRef]
- Bashir, A.; Klammer, A.; Robins, W.P.; Chin, C.-S.; Webster, D.; Paxinos, E.; Hsu, D.; Ashby, M.; Wang, S.; Peluso, P.; et al. A Hybrid Approach for the Automated Finishing of Bacterial Genomes. Nat. Biotechnol. 2012, 30, 701–707. [Google Scholar] [CrossRef]
- Wenger, A.M.; Peluso, P.; Rowell, W.J.; Chang, P.-C.; Hall, R.J.; Concepcion, G.T.; Ebler, J.; Fungtammasan, A.; Kolesnikov, A.; Olson, N.D.; et al. Accurate Circular Consensus Long-Read Sequencing Improves Variant Detection and Assembly of a Human Genome. Nat. Biotechnol. 2019, 37, 1155–1162. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, X.; Yekefenhazi, D.; Wang, J.; Zhong, K.; Zhang, Y.; Fu, H.; Zhou, Z.; Huang, J.; Li, W.; et al. Assembling Chromosome-Level Genomes of Male and Female Chanodichthys Mongolicus Using PacBio HiFi Reads and Hi-C Technologies. Sci. Data 2025, 12, 949. [Google Scholar] [CrossRef]
- Luo, Z.; Yi, M.; Yang, X.; Luo, Z.; Li, X.; Jiang, C.; Kang, B.; Huang, L.; Lin, H.-D.; He, X.; et al. The First High-Quality Chromosome-Level Genome of Parupeneus biaculeatus Using HiFi and Hi-C Data. Sci. Data 2025, 12, 1042. [Google Scholar] [CrossRef]
- Wick, R.R.; Holt, K.E. Benchmarking of Long-Read Assemblers for Prokaryote Whole Genome Sequencing. F1000Research 2021, 8, 2138. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Li, R.; Chen, C.; Sigwart, J.D.; Kocot, K.M. Benchmarking Oxford Nanopore Read Assemblers for High-Quality Molluscan Genomes. Philos. Trans. R. Soc. B Biol. Sci. 2021, 376, 20200160. [Google Scholar] [CrossRef] [PubMed]
- Espinosa, E.; Bautista, R.; Fernandez, I.; Larrosa, R.; Zapata, E.L.; Plata, O. Comparing Assembly Strategies for Third-Generation Sequencing Technologies across Different Genomes. Genomics 2023, 115, 110700. [Google Scholar] [CrossRef] [PubMed]
| Run1 | Run2 | Run3 | Run4 | Run5 | |
|---|---|---|---|---|---|
| sequencing depth | 40× | 31× | 24× | 34× | 24× |
| average read length | 8731.0 | 12,752.6 | 9516.5 | 9858.0 | 10,777.4 |
| read N50 | 11,283 | 17,814 | 12,208 | 15,869 | 15,519 |
| mean base quality | 14.4 | 14.0 | 13.9 | 19.2 | 17.2 |
| assembly length | 2,659,831,791 | 2,675,885,510 | 2,622,221,806 | 2,782,932,574 | 2,712,441,727 |
| longest contig | 48,309,394 | 61,879,406 | 14,254,794 | 24,617,929 | 39,123,270 |
| N50 | 11,819,014 | 14,961,077 | 2,831,721 | 5,079,353 | 7,100,512 |
| N75 | 5,723,742 | 7,086,134 | 1,627,221 | 2,449,746 | 3,320,268 |
| L50 | 70 | 54 | 290 | 160 | 119 |
| L75 | 150 | 119 | 591 | 358 | 252 |
| number of contigs | 5010 | 4687 | 6724 | 6963 | 8395 |
| Start | End | Coverage | n Contigs | L90 | L50 | Identity | n Blocks | |
|---|---|---|---|---|---|---|---|---|
| Chr 1 | 236 | 202,105,980 | 99.2 | 28 | 13 | 5 | 98.8 | 300 |
| Chr 2 | 1 | 188,946,972 | 98.6 | 33 | 16 | 6 | 99.0 | 294 |
| Chr 3 | 17,345 | 175,630,833 | 98.5 | 27 | 12 | 4 | 98.9 | 267 |
| Chr 4 | 17 | 165,320,435 | 98.5 | 39 | 17 | 6 | 99.0 | 298 |
| Chr 5 | 1 | 127,681,980 | 98.5 | 42 | 10 | 2 | 98.9 | 265 |
| Chr 6 | 1 | 120,552,326 | 98.8 | 34 | 14 | 5 | 99.0 | 177 |
| Chr 7 | 2174 | 117,119,118 | 98.9 | 16 | 9 | 3 | 98.9 | 173 |
| Chr 8 | 28,857 | 119,769,169 | 99.0 | 14 | 8 | 3 | 98.8 | 184 |
| Chr 9 | 1267 | 110,231,718 | 98.6 | 14 | 8 | 3 | 98.6 | 200 |
| Chr 10 | 3243 | 104,521,508 | 98.3 | 16 | 9 | 2 | 98.8 | 166 |
| Chr 11 | 1 | 102,289,349 | 98.2 | 21 | 12 | 4 | 99.0 | 154 |
| Chr 12 | 718 | 106,433,551 | 98.9 | 15 | 10 | 3 | 99.1 | 123 |
| Chr 13 | 16 | 90,494,031 | 96.5 | 54 | 13 | 4 | 98.9 | 202 |
| Chr 14 | 1994 | 83,494,928 | 99.2 | 33 | 15 | 6 | 99.0 | 127 |
| Chr 15 | 1 | 82,162,863 | 99.4 | 10 | 4 | 2 | 98.9 | 122 |
| Chr 16 | 4 | 84,651,008 | 96.9 | 24 | 8 | 2 | 98.8 | 166 |
| Chr 17 | 306 | 73,313,738 | 98.1 | 18 | 9 | 3 | 98.9 | 128 |
| Chr 18 | 1 | 65,914,046 | 97.9 | 21 | 8 | 3 | 99.0 | 123 |
| Chr 19 | 1 | 71,701,365 | 99.4 | 8 | 6 | 3 | 99.1 | 87 |
| Chr 20 | 5548 | 68,853,047 | 98.8 | 16 | 7 | 3 | 99.1 | 110 |
| Chr 21 | 1 | 60,856,787 | 98.6 | 9 | 6 | 3 | 99.0 | 97 |
| Chr 22 | 1 | 62,062,344 | 99.7 | 8 | 6 | 2 | 99.0 | 98 |
| Chr 23 | 20 | 51,730,624 | 98.9 | 17 | 5 | 2 | 98.8 | 97 |
| Chr 24 | 316 | 42,448,106 | 99.4 | 10 | 5 | 2 | 99.0 | 75 |
| Chr X | 8 | 143,533,377 | 92.6 | 308 | 196 | 46 | 99.3 | 490 |
| Run1 | Run2 | Run3 | Run4 | Run5 | |
|---|---|---|---|---|---|
| assembly length | 2,666,117,353 | 2,683,275,268 | 2,636,242,927 | 2,788,586,393 | 2,717,185,276 |
| N50 | 117,185,825 | 117,403,291 | 113,894,137 | 110,108,647 | 111,153,159 |
| N75 | 83,353,856 | 83,293,950 | 81,597,111 | 82,179,143 | 81,805,694 |
| L50 | 9 | 9 | 9 | 10 | 10 |
| L75 | 16 | 16 | 16 | 17 | 17 |
| n gaps | 1068 | 932 | 2090 | 1492 | 1166 |
| Start | End | Coverage | n Contigs | L90 | L50 | Identity | n Blocks | |
|---|---|---|---|---|---|---|---|---|
| Chr 1 | 236 | 202,105,980 | 99.2 | 6 | 1 | 1 | 98.8 | 296 |
| Chr 2 | 1 | 188,946,972 | 98.6 | 4 | 1 | 1 | 99.0 | 291 |
| Chr 3 | 17,345 | 175,630,833 | 98.5 | 6 | 1 | 1 | 98.9 | 270 |
| Chr 4 | 17 | 165,320,435 | 98.6 | 4 | 1 | 1 | 99.0 | 294 |
| Chr 5 | 1 | 127,681,980 | 98.5 | 12 | 1 | 1 | 98.9 | 260 |
| Chr 6 | 1 | 120,552,326 | 98.9 | 3 | 1 | 1 | 99.0 | 167 |
| Chr 7 | 2174 | 117,119,118 | 98.9 | 1 | 1 | 1 | 98.9 | 172 |
| Chr 8 | 28,857 | 119,769,169 | 98.9 | 1 | 1 | 1 | 98.8 | 180 |
| Chr 9 | 1267 | 110,231,718 | 98.7 | 1 | 1 | 1 | 98.6 | 201 |
| Chr 10 | 3243 | 104,521,508 | 98.2 | 1 | 1 | 1 | 98.8 | 162 |
| Chr 11 | 1 | 102,289,349 | 98.4 | 2 | 1 | 1 | 99.0 | 146 |
| Chr 12 | 718 | 106,433,551 | 98.9 | 1 | 1 | 1 | 99.1 | 124 |
| Chr 13 | 16 | 90,494,031 | 97.0 | 10 | 1 | 1 | 98.9 | 194 |
| Chr 14 | 1994 | 83,494,928 | 99.2 | 8 | 1 | 1 | 99.0 | 114 |
| Chr 15 | 1 | 82,162,863 | 99.4 | 1 | 1 | 1 | 98.9 | 118 |
| Chr 16 | 4 | 84,651,008 | 96.5 | 2 | 1 | 1 | 98.8 | 164 |
| Chr 17 | 306 | 73,313,738 | 98.1 | 6 | 1 | 1 | 98.9 | 122 |
| Chr 18 | 1 | 65,914,046 | 97.9 | 7 | 1 | 1 | 99.0 | 119 |
| Chr 19 | 1 | 71,701,365 | 99.4 | 1 | 1 | 1 | 99.1 | 82 |
| Chr 20 | 5548 | 68,853,047 | 98.9 | 3 | 1 | 1 | 99.1 | 106 |
| Chr 21 | 1 | 60,856,787 | 98.6 | 2 | 1 | 1 | 99.0 | 93 |
| Chr 22 | 1 | 62,062,344 | 99.7 | 1 | 1 | 1 | 99.0 | 93 |
| Chr 23 | 20 | 51,730,624 | 98.2 | 4 | 1 | 1 | 98.8 | 92 |
| Chr 24 | 316 | 42,448,106 | 99.4 | 1 | 1 | 1 | 99.0 | 74 |
| Chr X | 8 | 143,533,377 | 92.7 | 22 | 1 | 1 | 99.4 | 491 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toscano, E.; Sepe, L.; Di Maggio, F.; Nunziato, M.; Boccia, A.; Cimmino, E.; Scialla, A.; Salvatore, F.; Paolella, G. A Strategy for Single-Run Sequencing of the Water Buffalo Genome: (II) Fast One-Step Assembly of Highly Continuous Chromosome Sequences. Animals 2025, 15, 3014. https://doi.org/10.3390/ani15203014
Toscano E, Sepe L, Di Maggio F, Nunziato M, Boccia A, Cimmino E, Scialla A, Salvatore F, Paolella G. A Strategy for Single-Run Sequencing of the Water Buffalo Genome: (II) Fast One-Step Assembly of Highly Continuous Chromosome Sequences. Animals. 2025; 15(20):3014. https://doi.org/10.3390/ani15203014
Chicago/Turabian StyleToscano, Elvira, Leandra Sepe, Federica Di Maggio, Marcella Nunziato, Angelo Boccia, Elena Cimmino, Arcangelo Scialla, Francesco Salvatore, and Giovanni Paolella. 2025. "A Strategy for Single-Run Sequencing of the Water Buffalo Genome: (II) Fast One-Step Assembly of Highly Continuous Chromosome Sequences" Animals 15, no. 20: 3014. https://doi.org/10.3390/ani15203014
APA StyleToscano, E., Sepe, L., Di Maggio, F., Nunziato, M., Boccia, A., Cimmino, E., Scialla, A., Salvatore, F., & Paolella, G. (2025). A Strategy for Single-Run Sequencing of the Water Buffalo Genome: (II) Fast One-Step Assembly of Highly Continuous Chromosome Sequences. Animals, 15(20), 3014. https://doi.org/10.3390/ani15203014





