The Role of NFAT5 in Immune Response and Antioxidant Defense in the Thick-Shelled Mussel (Mytilus coruscus)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Full-Length Cloning and Bioinformatics Analysis of McNFAT5
2.3. Bacterial Challenge Experiment, RNA Extraction, and cDNA Synthesis
2.4. Quantitative Real-Time PCR Analysis
2.5. Immunohistochemistry
2.6. RNA Interference
2.7. Measurement of Antioxidant Capacity
2.8. Statistical Analysis
3. Results
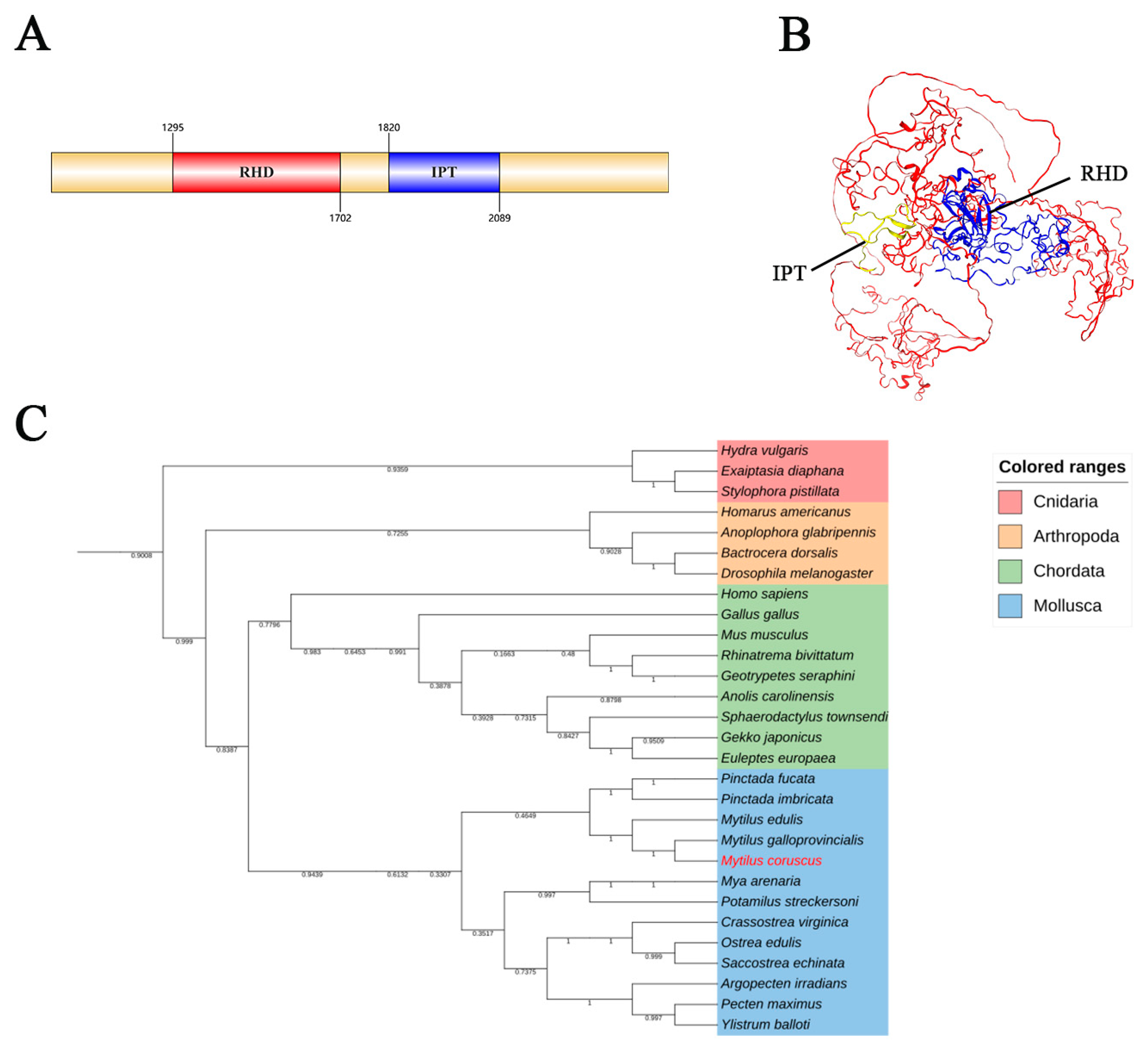
3.1. McNFAT5 Molecular Characterization
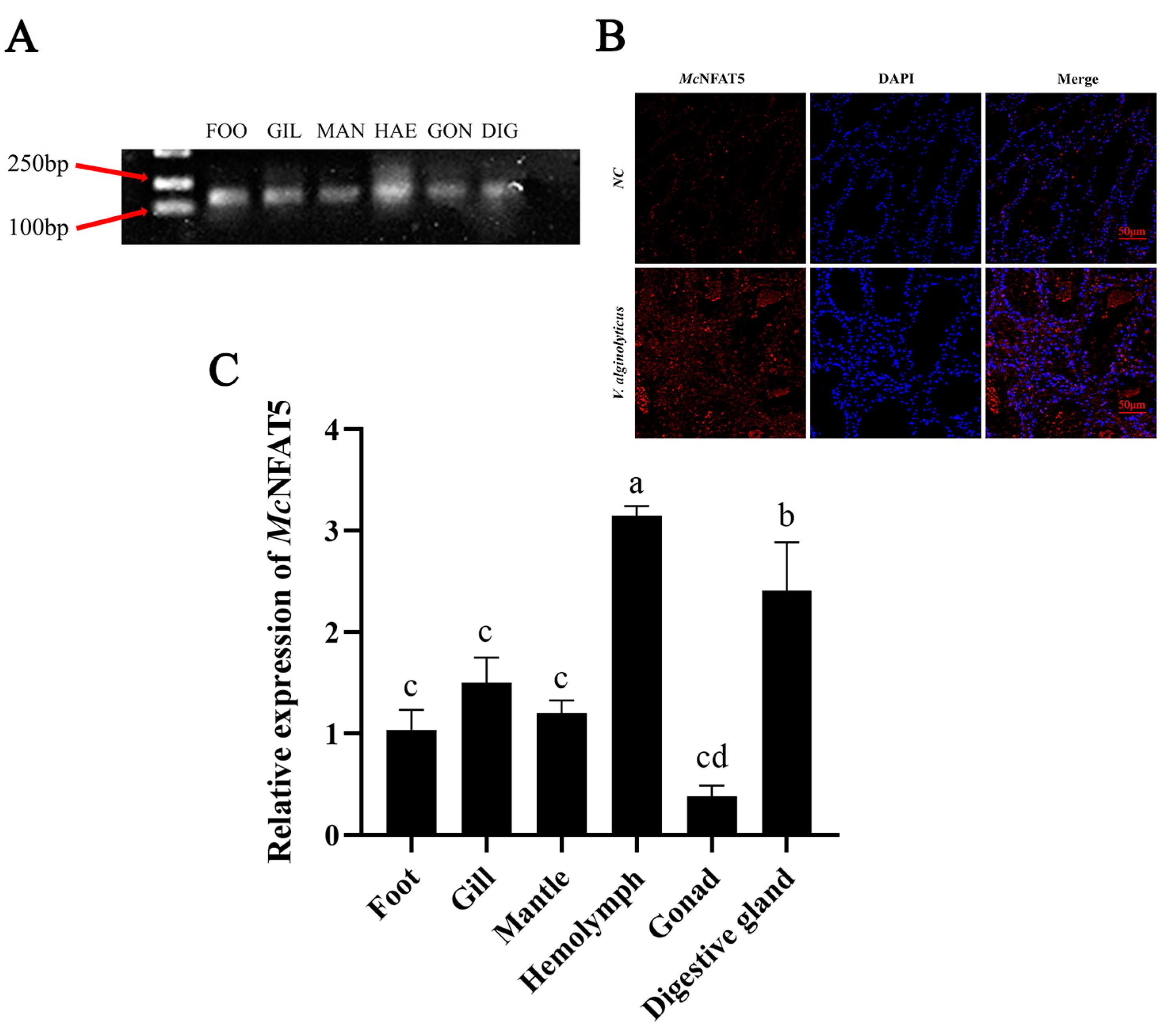
3.2. Differential McNFAT5 Gene Expression in Different Tissues After Pathogen Stimulation
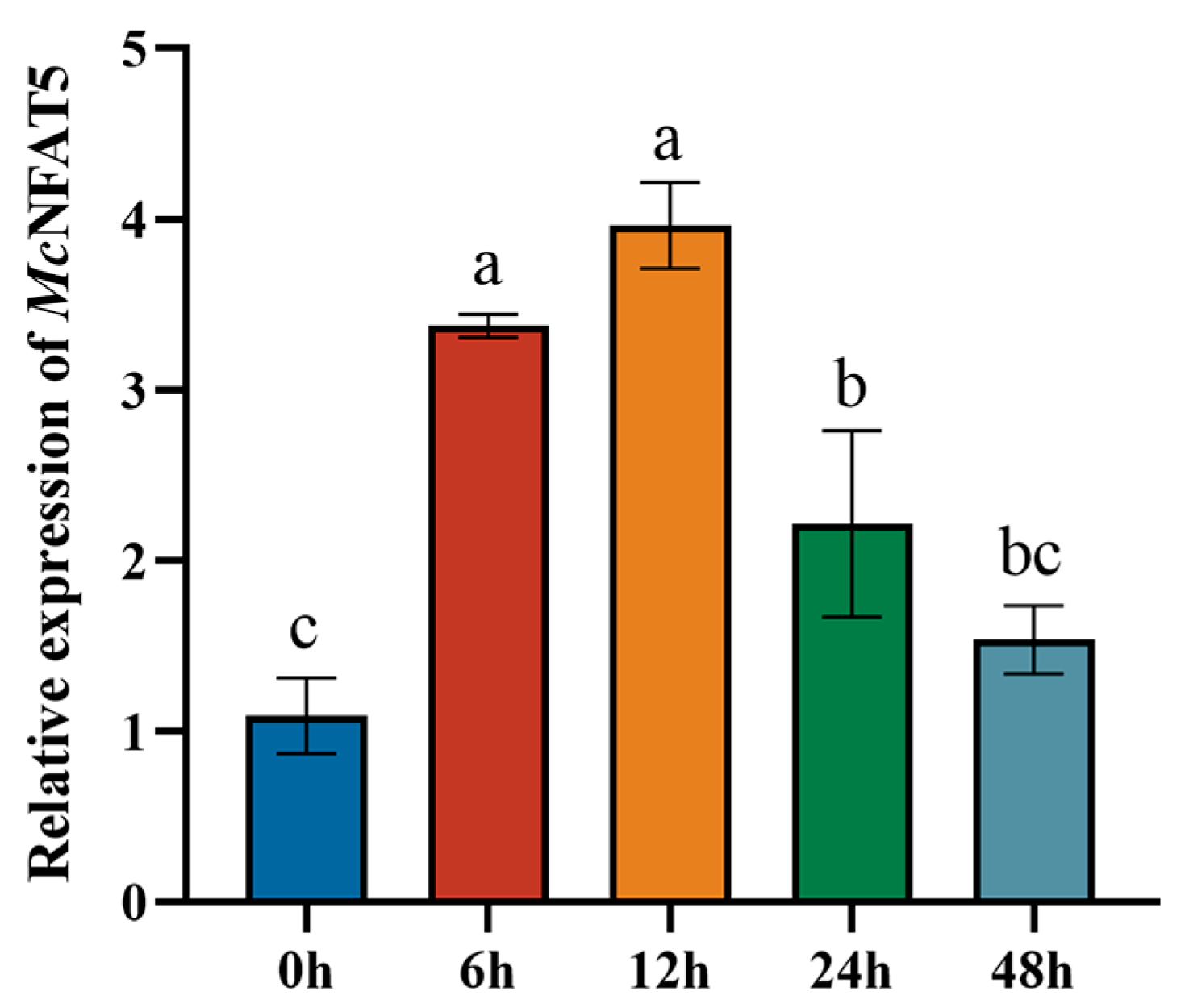
3.3. Temporal Expression Patterns of McNFAT5 After V. alginolyticus Stimulation
3.4. Effects of V. alginolyticus Infection on Antioxidants Activities
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dame, R.F.; Kenneth, M.J. Ecology of Marine Bivalves: An Ecosystem Approach; Taylor & Francis: Abingdon, UK, 2011. [Google Scholar]
- Zannella, C.; Mosca, F.; Mariani, F.; Franci, G.; Folliero, V.; Galdiero, M.; Tiscar, P.G.; Galdiero, M. Microbial diseases of bivalve mollusks: Infections, immunology and antimicrobial defense. Mar. Drugs 2017, 15, 182. [Google Scholar] [CrossRef]
- Shaw, J.-P.; Utz, P.J.; Durand, D.B.; Toole, J.J.; Emmel, E.A.; Crabtree, G.R. Identification of a putative regulator of early T cell activation genes. Science 1988, 241, 202–205. [Google Scholar] [CrossRef] [PubMed]
- Macian, F. NFAT proteins: Key regulators of T-cell development and function. Nat. Rev. Immunol. 2005, 5, 472–484. [Google Scholar] [CrossRef]
- Fric, J.; Zelante, T.; Wong, A.Y.; Mertes, A.; Yu, H.-B.; Ricciardi-Castagnoli, P. NFAT control of innate immunity. J. Am. Soc. Hematol. 2012, 120, 1380–1389. [Google Scholar] [CrossRef] [PubMed]
- Zanoni, I.; Granucci, F. Regulation and dysregulation of innate immunity by NFAT signaling downstream of pattern recognition receptors (PRRs). Eur. J. Immunol. 2012, 42, 1924–1931. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-U.; Kim, L.-K.; Choi, J.-M. Revisiting the concept of targeting NFAT to control T cell immunity and autoimmune diseases. Front. Immunol. 2018, 9, 2747. [Google Scholar] [CrossRef]
- Hogan, P.G.; Chen, L.; Nardone, J.; Rao, A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 2003, 17, 2205–2232. [Google Scholar] [CrossRef]
- Trama, J.P. Identification and Characterization of NFAT5: A Novel Rel Family Transcription Factor; University of California, San Diego: San Diego, CA, USA, 2002. [Google Scholar]
- Graef, I.A.; Gastier, J.M.; Francke, U.; Crabtree, G.R. Evolutionary relationships among Rel domains indicate functional diversification by recombination. Proc. Natl. Acad. Sci. USA 2001, 98, 5740–5745. [Google Scholar] [CrossRef]
- Sullivan, J.C.; Kalaitzidis, D.; Gilmore, T.D.; Finnerty, J.R. Rel homology domain-containing transcription factors in the cnidarian Nematostella vectensis. Dev. Genes Evol. 2007, 217, 63–72. [Google Scholar] [CrossRef]
- Berga-Bolanos, R.; Drews-Elger, K.; Aramburu, J.; Lopez-Rodriguez, C. NFAT5 regulates T lymphocyte homeostasis and CD24-dependent T cell expansion under pathologic hypernatremia. J. Immunol. 2010, 185, 6624–6635. [Google Scholar] [CrossRef]
- Tellechea, M.; Buxadé, M.; Tejedor, S.; Aramburu, J.; Lopez-Rodriguez, C. NFAT5-regulated macrophage polarization supports the proinflammatory function of macrophages and T lymphocytes. J. Immunol. 2018, 200, 305–315. [Google Scholar] [CrossRef]
- Ko, B.C.; Lam, A.K.; Kapus, A.; Fan, L.; Chung, S.K.; Chung, S.S. Fyn and p38 signaling are both required for maximal hypertonic activation of the osmotic response element-binding protein/tonicity-responsive enhancer-binding protein (OREBP/TonEBP). J. Biol. Chem. 2002, 277, 46085–46092. [Google Scholar] [CrossRef]
- Song, X.; Hu, J.; Jin, P.; Chen, L.; Ma, F. Identification and evolution of an NFAT gene involving Branchiostoma belcheri innate immunity. Genomics 2013, 102, 355–362. [Google Scholar] [CrossRef]
- Letunic, I.; Doerks, T.; Bork, P. SMART: Recent updates, new developments and status in 2015. Nucleic Acids Res. 2015, 43, D257–D260. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Qi, P.; Wu, Y.; Gu, Z.; Li, H.; Li, J.; Guo, B.; Liao, Z.; Yan, X. A novel molluscan TLR molecule engaged in inflammatory response through MyD88 adapter recruitment. Dev. Comp. Immunol. 2022, 131, 104373. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Li, Z.; Chang, X.; Hu, M.; Fang, J.K.-H.; Sokolova, I.M.; Huang, W.; Xu, E.G.; Wang, Y. Is microplastic an oxidative stressor? Evidence from a meta-analysis on bivalves. J. Hazard. Mater. 2022, 423, 127211. [Google Scholar] [CrossRef]
- Jia, S.; Flores-Saaib, R.n.D.; Courey, A.J. The Dorsal Rel homology domain plays an active role in transcriptional regulation. Mol. Cell. Biol. 2002, 22, 5089–5099. [Google Scholar] [CrossRef]
- Huang, T. Study on the Tonicity-Regulated Nucleocytoplasmic Trafficking of NT5 Using Proteomics and Functional siRNA Screen. Ph.D. Thesis, The Chinese University of Hong Kong, Hong Kong, China, 2015. [Google Scholar]
- Huang, X.-D.; Wei, G.-j.; Zhang, H.; He, M.-X. Nuclear factor of activated T cells (NFAT) in pearl oyster Pinctada fucata: Molecular cloning and functional characterization. Fish Shellfish. Immunol. 2015, 42, 108–113. [Google Scholar] [CrossRef]
- Duan, X.; Lv, M.; Liu, A.; Pang, Y.; Li, Q.; Su, P.; Gou, M. Identification and evolution of transcription factors RHR gene family (NFAT and RBPJ) involving lamprey (Lethenteron reissneri) innate immunity. Mol. Immunol. 2021, 138, 38–47. [Google Scholar] [CrossRef]
- Feng, Z.-P. An overview on predicting the subcellular location of a protein. Silico Biol. 2002, 2, 291–303. [Google Scholar]
- Marafiot, T.; Pozzobon, M.; Hansmann, M.L.; Ventura, R.; Pileri, S.A.; Roberton, H.; Gesk, S.; Gaulard, P.; Barth, T.F.; Du, M.Q. The NFATc1 transcription factor is widely expressed in white cells and translocates from the cytoplasm to the nucleus in a subset of human lymphomas. Br. J. Haematol. 2005, 128, 333–342. [Google Scholar] [CrossRef]
- Silva, R.C.M.C.; Gomes, F.M. Evolution of the Major Components of Innate Immunity in Animals. J. Mol. Evol. 2024, 92, 3–20. [Google Scholar] [CrossRef]
- Kim, N.H.; Choi, S.; Han, E.J.; Hong, B.K.; Choi, S.Y.; Kwon, H.M.; Hwang, S.Y.; Cho, C.S.; Kim, W.U. The xanthine oxidase–NFAT5 pathway regulates macrophage activation and TLR-induced inflammatory arthritis. Eur. J. Immunol. 2014, 44, 2721–2736. [Google Scholar] [CrossRef]
- Lee, J.H.; Suh, J.H.; Choi, S.Y.; Kang, H.J.; Lee, H.H.; Ye, B.J.; Lee, G.R.; Jung, S.W.; Kim, C.J.; Lee-Kwon, W. Tonicity-responsive enhancer-binding protein promotes hepatocellular carcinogenesis, recurrence and metastasis. Gut 2019, 68, 347–358. [Google Scholar] [CrossRef]
- Chaitanya, R.; Shashank, K.; Sridevi, P. Oxidative stress in invertebrate systems. Free. Radicla Dis. 2016, 19, 51–68. [Google Scholar]
- Sahu, P.K.; Jayalakshmi, K.; Tilgam, J.; Gupta, A.; Nagaraju, Y.; Kumar, A.; Hamid, S.; Singh, H.V.; Minkina, T.; Rajput, V.D. ROS generated from biotic stress: Effects on plants and alleviation by endophytic microbes. Front. Plant Sci. 2022, 13, 1042936. [Google Scholar] [CrossRef]
- Hollborn, M.; Fischer, S.; Kuhrt, H.; Wiedemann, P.; Bringmann, A.; Kohen, L. Osmotic regulation of NFAT5 expression in RPE cells: The involvement of purinergic receptor signaling. Mol. Vis. 2017, 23, 116. [Google Scholar]
- Ighodaro, O.; Akinloye, O.A. First line defence antioxidants-superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPX): Their fundamental role in the entire antioxidant defence grid. Alex. J. Med. 2018, 54, 287–293. [Google Scholar] [CrossRef]

| Primer | Sequences (5′–3′) | Usage |
|---|---|---|
| McNFAT5-F | TCTTCTTGACCGTGCTGGAC | qRT–PCR |
| McNFAT5-R | TCGTCGGACTTTTGGCACTT | |
| McTRAF-F | TGTGCCAATTCCCTGTCCT | qRT–PCR |
| McTRAF-R | GGACACTCTTTATGCAGG | |
| McIRAK-F | CCTTTTATGGCAGCAGCGTG | qRT–PCR |
| McIRAK-R | AAAATCCAGTGCCCGATGGT | |
| Mcmyticofensin-F | TGTGGCTCTAGAAGTTGCTGATG | qRT–PCR |
| Mc myticofensin-R | TCAATCTGAACCAGCCTCCAC | |
| β-actin-F | GCTACGAATTACCTGACGGAC | qRT–PCR |
| β-actin-R | TTCCCAAGAAAGATGGTTGTAACAT | |
| siNC | UUCUCCGAACGUGUCACGUTT | RNAi |
| ACGUGACACGUUCGGAGAATT | ||
| SiNFAT5 | GACAAUAAAUCAACUGUUATT | RNAi |
| UAACAGUUGAUUUAUUGUCTT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bei, Y.; Si, X.; Ma, W.; Qi, P.; Ye, Y. The Role of NFAT5 in Immune Response and Antioxidant Defense in the Thick-Shelled Mussel (Mytilus coruscus). Animals 2025, 15, 726. https://doi.org/10.3390/ani15050726
Bei Y, Si X, Ma W, Qi P, Ye Y. The Role of NFAT5 in Immune Response and Antioxidant Defense in the Thick-Shelled Mussel (Mytilus coruscus). Animals. 2025; 15(5):726. https://doi.org/10.3390/ani15050726
Chicago/Turabian StyleBei, Yijiang, Xirui Si, Wenjun Ma, Pengzhi Qi, and Yingying Ye. 2025. "The Role of NFAT5 in Immune Response and Antioxidant Defense in the Thick-Shelled Mussel (Mytilus coruscus)" Animals 15, no. 5: 726. https://doi.org/10.3390/ani15050726
APA StyleBei, Y., Si, X., Ma, W., Qi, P., & Ye, Y. (2025). The Role of NFAT5 in Immune Response and Antioxidant Defense in the Thick-Shelled Mussel (Mytilus coruscus). Animals, 15(5), 726. https://doi.org/10.3390/ani15050726






