Study on the Regulatory Mechanisms of Carapace Marking Formation in Marsupenaeus japonicus
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. RNA Extraction and cDNA Synthesis
2.3. Construction and Sequencing of the miRNA Library
2.4. miRNA Bioinformatics Analysis
2.5. Differentially Expressed miRNA Analysis and miRNA Target Prediction
2.6. Validation of miRNA Expression by Real-Time Quantitative PCR
2.7. Tissue Expression Characteristics of Crustacyanin Genes
2.8. RNAi Experiment of Crustacyanin Genes
3. Results
3.1. Analysis and Identification of Small RNAs
3.2. Differentially Expressed miRNA Analysis
3.3. miRNA Target Gene Prediction and Functional Analysis
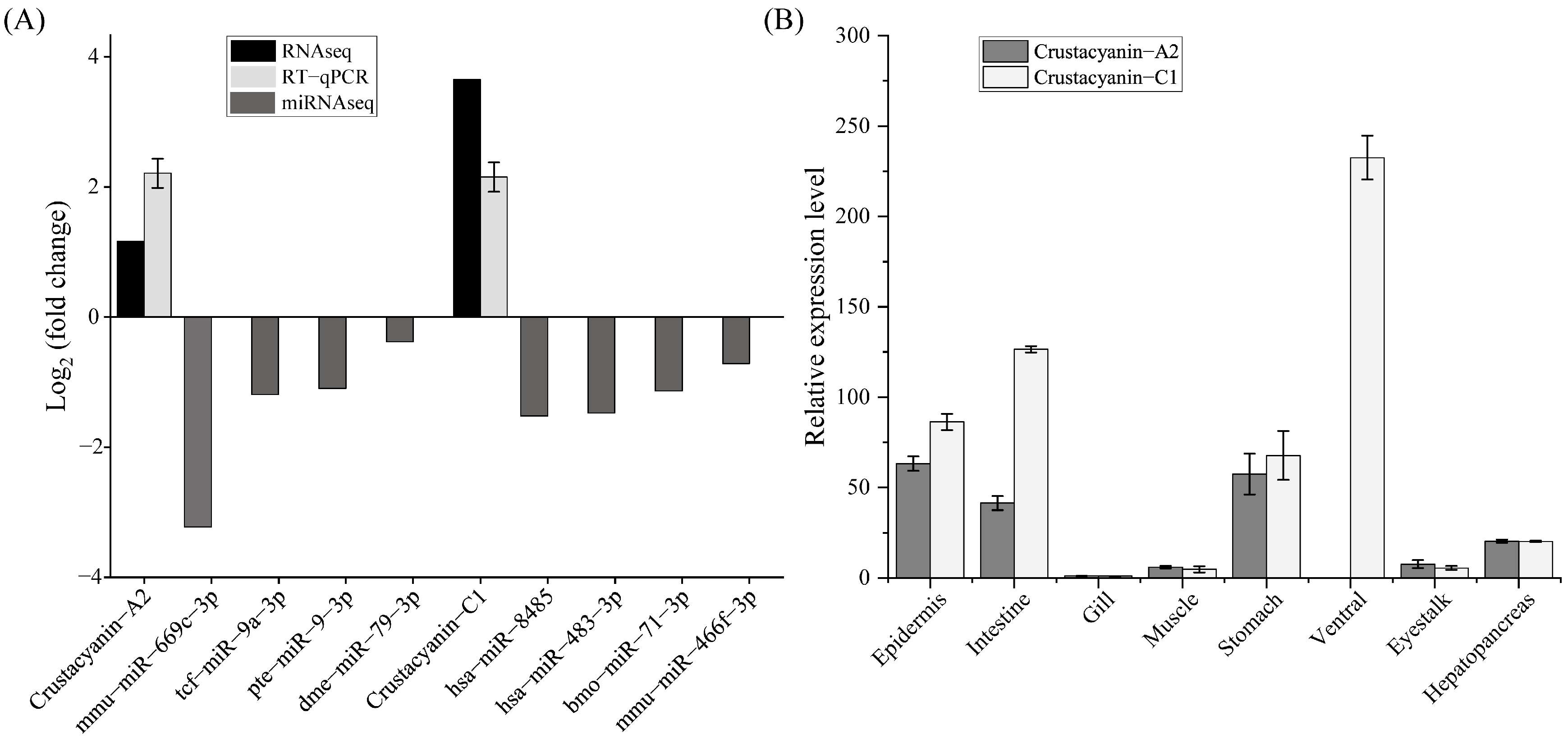
3.4. Verification of Differential miRNA by qRT-PCR
3.5. The Expression Characteristics of Crustacyanin Genes
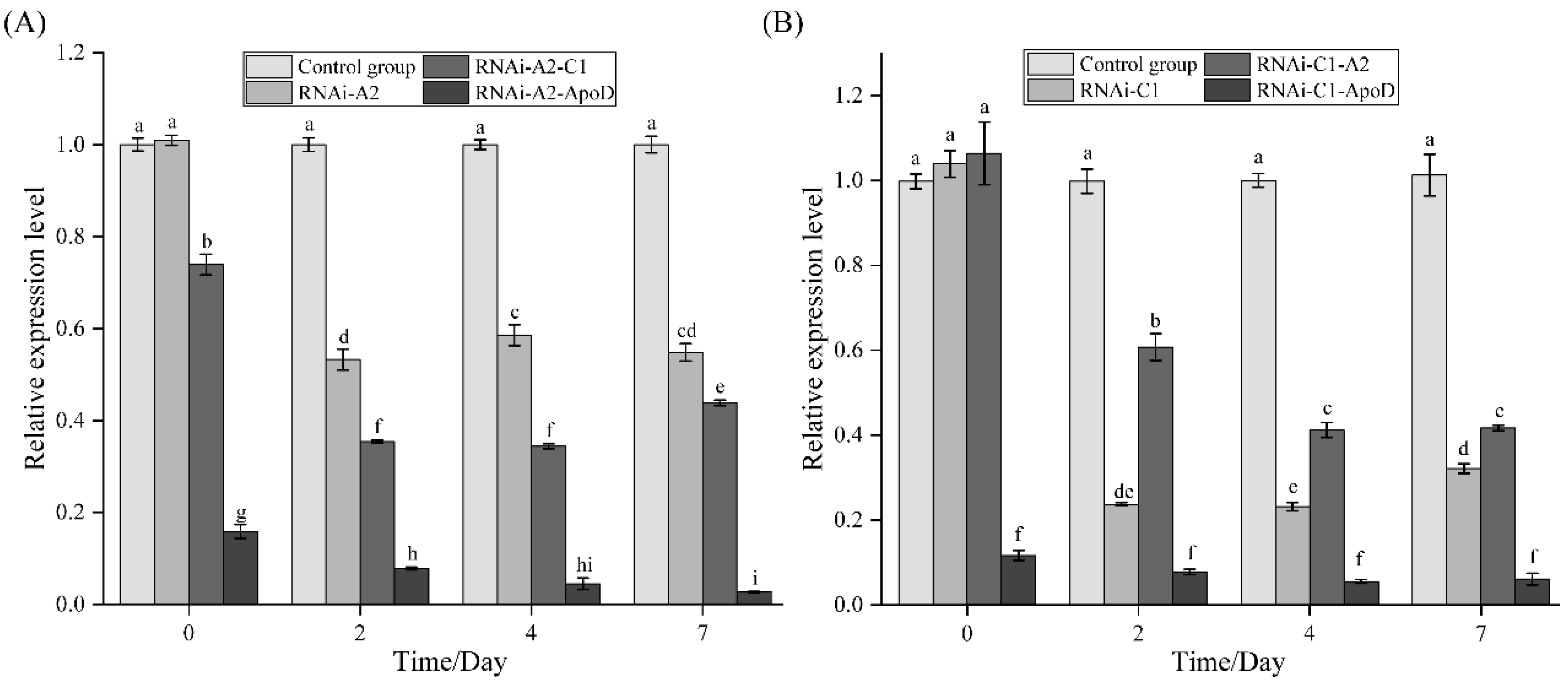
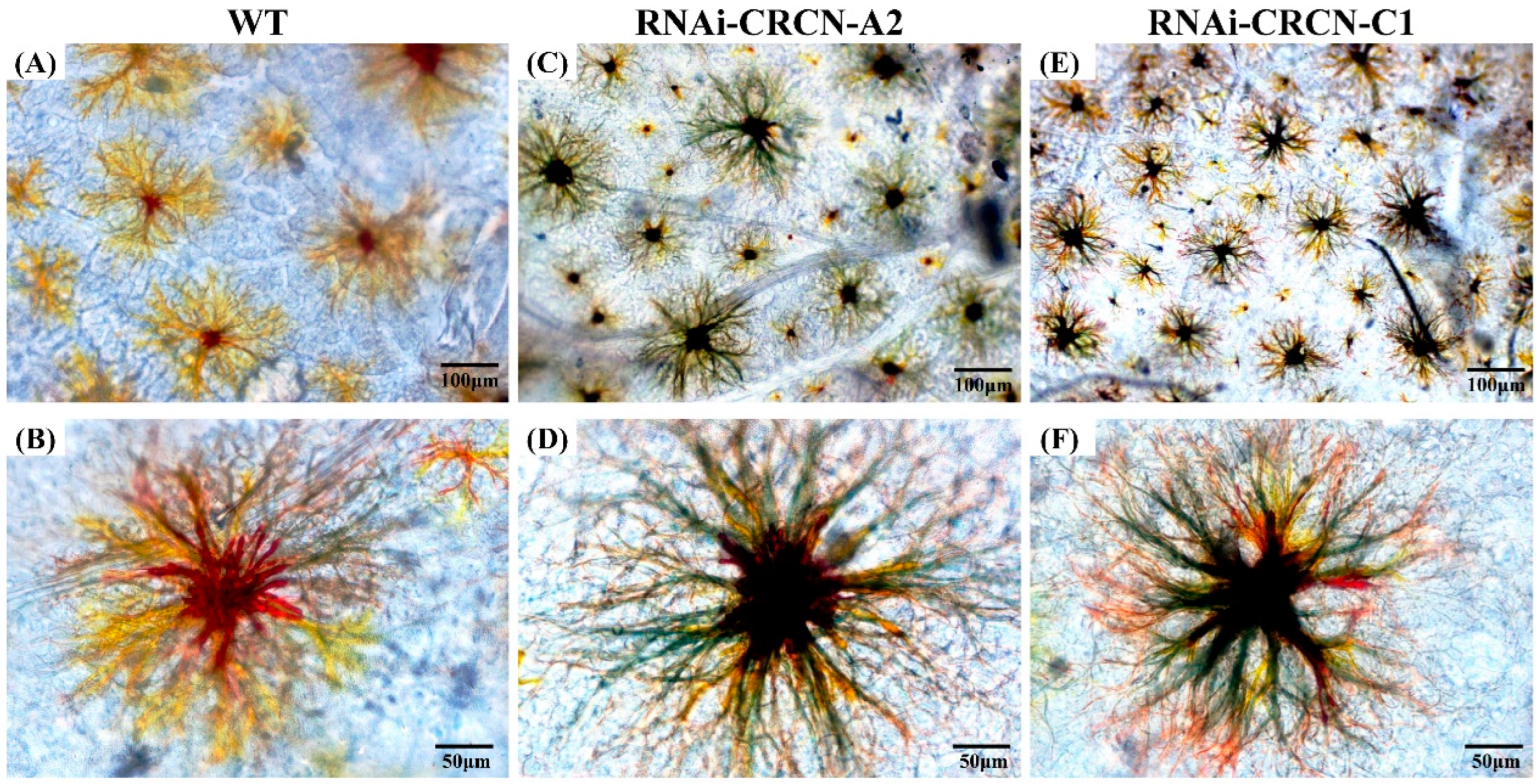
3.6. Effects of Crustacyanin Gene Silencing on the Expression of Related Genes and Chromatophores of the Carapace
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pérez Farfante, I.; Kensley, B. Penaeoid and sergestoid shrimps and prawns of the world. Keys and diagnoses for the families and genera. Mémoires Du Muséum Natl. D’histoire Nat. 1997, 175, 1–233. [Google Scholar]
- Tsoi, K.; Wang, Z.; Chu, K. Genetic divergence between two morphologically similar varieties of the kuruma shrimp Penaeus japonicus. Mar. Biol. 2005, 147, 367–379. [Google Scholar] [CrossRef]
- Tsoi, K.; Chan, T.-Y.; Chu, K. Molecular population structure of the kuruma shrimp Penaeus japonicus species complex in western Pacific. Mar. Biol. 2007, 150, 1345–1364. [Google Scholar] [CrossRef]
- Tsoi, K.H.; Ma, K.Y.; Wu, T.; Fennessy, S.T.; Chu, K.H.; Chan, T.Y. Verification of the cryptic species Penaeus pulchricaudatus in the commercially important kuruma shrimp P. japonicus (Decapoda: Penaeidae) using molecular taxonomy. Invertebr. Syst. 2014, 28, 476–490. [Google Scholar] [CrossRef]
- Wang, P.; Xie, S.; Li, X.; Zhu, J.; You, Z.; Zhou, X.; Zhou, X.; Yan, B.; Gao, H.; Xing, C. Transcriptome analysis provides insights into the mechanism of carapace stripe formation in two closely related Marsupenaeus species. Front. Mar. Sci. 2023, 10, 1234940. [Google Scholar] [CrossRef]
- Border, S.E.; Piefke, T.J.; Fialkowski, R.J.; Tryc, M.R.; Funnell, T.R.; DeOliveira, G.M.; Dijkstra, P.D. Color change and pigmentation in a color polymorphic cichlid fish. Hydrobiologia 2019, 832, 175–191. [Google Scholar] [CrossRef]
- Vissio, P.G.; Darias, M.J.; Di Yorio, M.P.; Sirkin, D.I.P.; Delgadin, T.H. Fish skin pigmentation in aquaculture: The influence of rearing conditions and its neuroendocrine regulation. Gen. Comp. Endocrinol. 2021, 301, 113662. [Google Scholar] [CrossRef] [PubMed]
- Lemoine, M.; Barluenga, M.; Lucek, K.; Mwaiko, S.; Haesler, M.; Chapman, L.J.; Chapman, C.A.; Seehausen, O. Recent sympatric speciation involving habitat-associated nuptial colour polymorphism in a crater lake cichlid. Hydrobiologia 2019, 832, 297–315. [Google Scholar] [CrossRef]
- Berdan, E.L.; Fuller, R.C.; Kozak, G.M. Genomic landscape of reproductive isolation in Lucania killifish: The role of sex loci and salinity. J. Evol. Biol. 2021, 34, 157–174. [Google Scholar] [CrossRef]
- Hao, R.; Zhu, X.; Tian, C.; Jiang, M.; Huang, Y.; Zhu, C. Integrated analysis of the role of miRNA-mRNA in determining different body colors of leopard coral grouper (Plectropomus leopardus). Aquaculture 2022, 548, 737575. [Google Scholar] [CrossRef]
- Yan, B.; Liu, B.; Zhu, C.-D.; Li, K.-L.; Yue, L.-J.; Zhao, J.-L.; Gong, X.-L.; Wang, C.-H. microRNA regulation of skin pigmentation in fish. J. Cell Sci. 2013, 126, 3401–3408. [Google Scholar]
- Cai, Y.; Yu, X.; Hu, S.; Yu, J. A brief review on the mechanisms of miRNA regulation. Genom. Proteom. Bioinform. 2009, 7, 147–154. [Google Scholar] [CrossRef]
- Wongdontri, C.; Luangtrakul, W.; Boonchuen, P.; Sarnow, P.; Somboonviwat, K.; Jaree, P.; Somboonwiwat, K. Participation of shrimp pva-miR-166 in hemocyte homeostasis by modulating apoptosis-related gene PvProsaposin during white spot syndrome virus infection. J. Virol. 2024, 98, e0053024. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Pang, X.; Wang, L.; Li, M.; Dong, C.; Ma, X.; Wang, L.; Song, D.; Feng, J.; Xu, P. Dynamic regulation of mRNA and miRNA associated with the developmental stages of skin pigmentation in Japanese ornamental carp. Gene 2018, 666, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Lu, Z.-C.; Zhu, X.-W.; Zhu, C.-H.; Wang, C.-G.; Shen, Y.-C.; Wang, W. Differential expression of microRNAs in hemocytes from white shrimp Litopenaeus vannamei under copper stress. Fish Shellfish Immunol. 2018, 74, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Luo, M.; Jiang, B.; Zhu, W.; Min, Q.; Hu, J.; Liu, T.; Fu, J.; Shi, X.; Wang, P. microRNA regulation of skin pigmentation in golden-back mutant of crucian carp from a rice-fish integrated farming system. BMC Genom. 2023, 24, 70. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Wang, L.; Zhu, W.; Fu, J.; Song, F.; Fang, M.; Dong, J.; Dong, Z. Identification and characterization of skin color microRNAs in koi carp (Cyprinus carpio L.) by Illumina sequencing. BMC Genom. 2018, 19, 779. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Luo, M.; Luo, W.; Wang, L.; Zhu, W.; Fu, J.; Dong, Z. miR-196a regulates the skin pigmentation of koi carp (Cyprinus carpio L.) by targeting transcription factor mitfa. Aquac. Res. 2021, 52, 229–236. [Google Scholar] [CrossRef]
- Dong, Z.; Luo, M.; Wang, L.; Yin, H.; Zhu, W.; Fu, J. MicroRNA-206 regulation of skin pigmentation in koi carp (Cyprinus carpio L.). Front. Genet. 2020, 11, 47. [Google Scholar] [CrossRef]
- Wu, S.; Huang, J.; Li, Y.; Zhao, L. Involvement of miR-495 in the skin pigmentation of rainbow trout (Oncorhynchus mykiss) through the regulation of mc1r. Int. J. Biol. Macromol. 2024, 254, 127638. [Google Scholar] [CrossRef] [PubMed]
- Angell, A.; de Nys, R.; Mangott, A.; Vucko, M.J. The effects of concentration and supplementation time of natural and synthetic sources of astaxanthin on the colouration of the prawn Penaeus monodon. Algal Res. 2018, 35, 577–585. [Google Scholar] [CrossRef]
- Jin, Y.; Li, S.; Yu, Y.; Zhang, C.; Zhang, X.; Li, F. Transcriptome Analysis Provides Insights into the Mechanism of Astaxanthin Enrichment in a Mutant of the Ridgetail White Prawn Exopalaemon carinicauda. Genes 2021, 12, 618. [Google Scholar] [CrossRef] [PubMed]
- Maoka, T.; Kawashima, Y.; Takaki, M. Structures of yellow xanthophylls and metabolism of astaxanthin in the prawn Penaeus japonicus. J. Oleo Sci. 2018, 67, 1425–1433. [Google Scholar] [CrossRef]
- Tan, K.; Zhang, H.; Lim, L.-S.; Ma, H.; Li, S.; Zheng, H. Roles of carotenoids in invertebrate immunology. Front. Immunol. 2020, 10, 3041. [Google Scholar] [CrossRef]
- Maoka, T. Carotenoids as natural functional pigments. J. Nat. Med. 2020, 74, 1–16. [Google Scholar] [CrossRef]
- Wade, N.M.; Gabaudan, J.; Glencross, B.D. A review of carotenoid utilisation and function in crustacean aquaculture. Rev. Aquac. 2017, 9, 141–156. [Google Scholar] [CrossRef]
- Begum, S.; Cianci, M.; Durbeej, B.; Falklöf, O.; Hädener, A.; Helliwell, J.R.; Helliwell, M.; Regan, A.C.; Watt, C.I.F. On the origin and variation of colors in lobster carapace. Phys. Chem. Chem. Phys. 2015, 17, 16723–16732. [Google Scholar] [CrossRef]
- Chen, H.; Ji, H.; Pan, C.; Zhang, D.; Su, W.; Liu, S.; Deng, Y.; Huang, X. Purification and Characterisation of Two Novel Pigment Proteins from the Carapace of Red Swamp Crayfish (Procambarus clarkii). Foods 2022, 11, 35. [Google Scholar] [CrossRef]
- Zhao, C.; Peng, C.; Fan, S.; Wang, P.; Yan, L.; Xie, Z.; Qiu, L. Identification of a novel crustacyanin-like lipocalin in Penaeus monodon: Molecular cloning, tissue distribution and its functional studies in astaxanthin accumulation. Aquaculture 2021, 539, 736615. [Google Scholar] [CrossRef]
- Wade, N.M.; Anderson, M.; Sellars, M.J.; Tume, R.K.; Preston, N.P.; Glencross, B.D. Mechanisms of colour adaptation in the prawn Penaeus monodon. J. Exp. Biol. 2012, 215, 343–350. [Google Scholar] [CrossRef]
- Wade, N.M.; Tollenaere, A.; Hall, M.R.; Degnan, B.M. Evolution of a novel carotenoid-binding protein responsible for crustacean shell color. Mol. Biol. Evol. 2009, 26, 1851–1864. [Google Scholar] [CrossRef]
- Budd, A.M.; Hinton, T.M.; Tonks, M.; Cheers, S.; Wade, N.M. Rapid expansion of pigmentation genes in penaeid shrimp with absolute preservation of function. J. Exp. Biol. 2017, 220, 4109–4118. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Ma, H.; Sun, J.; Xu, W.; Gao, W.; Lai, X.; Yan, B. Expression and function analysis of crustacyanin gene family involved in resistance to heavy metal stress and body color formation in Exopalaemon carinicauda. J. Exp. Zool. Part B Mol. Dev. Evol. 2021, 336, 352–363. [Google Scholar] [CrossRef]
- Yang, F.; Wang, M.R.; Ma, Y.G.; Ma, W.M.; Yang, W.J. Prawn lipocalin: Characterization of a color shift induced by gene knockdown and ligand binding assay. J. Exp. Zool. Part A Ecol. Genet. Physiol. 2011, 315, 562–571. [Google Scholar] [CrossRef] [PubMed]
- Ertl, N.G.; Elizur, A.; Brooks, P.; Kuballa, A.V.; Anderson, T.A.; Knibb, W.R. Molecular characterisation of colour formation in the prawn Fenneropenaeus merguiensis. PLoS ONE 2013, 8, e56920. [Google Scholar] [CrossRef]
- von Lintig, J.; Moon, J.; Lee, J.; Ramkumar, S. Carotenoid metabolism at the intestinal barrier. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2020, 1865, 158580. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zhang, L.; Wang, G.; Huang, S. De novo assembly transcriptome analysis reveals the genes associated with body color formation in the freshwater ornamental shrimps Neocaridina denticulate sinensis. Gene 2022, 806, 145929. [Google Scholar] [CrossRef]
- Bhosale, P.; Bernstein, P.S. Vertebrate and invertebrate carotenoid-binding proteins. Arch. Biochem. Biophys. 2007, 458, 121–127. [Google Scholar] [CrossRef]
- Lin, S.; Zhang, L.; Wang, G.; Huang, S.; Wang, Y. Searching and identifying pigmentation genes from Neocaridina denticulate sinensis via comparison of transcriptome in different color strains. Comp. Biochem. Physiol. Part D Genom. Proteom. 2022, 42, 100977. [Google Scholar] [CrossRef]
- Griffiths-Jones, S. miRBase: microRNA sequences and annotation. Curr. Protoc. Bioinform. 2010, 29, 12.9.1–12.9.10. [Google Scholar] [CrossRef]
- Friedländer, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Wen, M.; Shen, Y.; Shi, S.; Tang, T. miREvo: An integrative microRNA evolutionary analysis platform for next-generation sequencing experiments. BMC Bioinform. 2012, 13, 1–10. [Google Scholar] [CrossRef]
- Zhou, L.; Chen, J.; Li, Z.; Li, X.; Hu, X.; Huang, Y.; Zhao, X.; Liang, C.; Wang, Y.; Sun, L. Integrated profiling of microRNAs and mRNAs: microRNAs located on Xq27. 3 associate with clear cell renal cell carcinoma. PLoS ONE 2010, 5, e15224. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Enright, A.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D. MicroRNA targets in Drosophila. Genome Biol. 2003, 5, R1. [Google Scholar] [CrossRef]
- Krüger, J.; Rehmsmeier, M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006, 34, W451–W454. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. goseq: Gene Ontology testing for RNA-seq datasets. R Bioconductor 2012, 8, 1–25. [Google Scholar]
- Bedini, R.; Canali, M.G.; Bedini, A. Functions of the eyespot marking on the swimming leg of Liocarcinus depurator (Linnaeus, 1758) (Decapoda, Brachyura, Portunidae). Crustaceana 2002, 75, 979–992. [Google Scholar] [CrossRef]
- McNamara, J.C.; Milograna, S.R. Adaptive color change and the molecular endocrinology of pigment translocation in crustacean chromatophores. Nat. Hist. Crustac. 2015, 4, 68–102. [Google Scholar]
- Watanabe, M.; Kondo, S. Is pigment patterning in fish skin determined by the Turing mechanism? Trends Genet. 2015, 31, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Kelsh, R.N. Genetics and evolution of pigment patterns in fish. Pigment Cell Res. 2004, 17, 326–336. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zhang, S.; Xu, J.; Feng, J.; Mahboob, S.; Al-Ghanim, K.A.; Sun, X.; Xu, P. Comparative transcriptome analysis reveals the genetic basis of skin color variation in common carp. PLoS ONE 2014, 9, e108200. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Stricker, H.M.; Gou, D.; Liu, L. MicroRNA: Past and present. Front Biosci 2007, 12, C2329. [Google Scholar] [CrossRef] [PubMed]
- Chiang, H.R.; Schoenfeld, L.W.; Ruby, J.G.; Auyeung, V.C.; Spies, N.; Baek, D.; Johnston, W.K.; Russ, C.; Luo, S.; Babiarz, J.E. Mammalian microRNAs: Experimental evaluation of novel and previously annotated genes. Genes Dev. 2010, 24, 992–1009. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Trombly, M.I.; Chen, J.; Wang, X. Essential and overlapping functions for mammalian Argonautes in microRNA silencing. Genes Dev. 2009, 23, 304–317. [Google Scholar] [CrossRef]
- Li, X.; Ponandai-Srinivasan, S.; Nandakumar, K.S.; Fabre, S.; Xu Landén, N.; Mavon, A.; Khmaladze, I. Targeting microRNA for improved skin health. Health Sci. Rep. 2021, 4, e374. [Google Scholar] [CrossRef]
- Feng, D.; Li, Q.; Yu, H.; Liu, S.; Kong, L.; Du, S. Integrated analysis of microRNA and mRNA expression profiles in Crassostrea gigas to reveal functional miRNA and miRNA-targets regulating shell pigmentation. Sci. Rep. 2020, 10, 20238. [Google Scholar] [CrossRef]
- Zheng, J.; Cao, J.; Mao, Y.; Su, Y.; Wang, J. Identification of microRNAs with heat stress responsive and immune properties in Marsupenaeus japonicus based on next-generation sequencing and bioinformatics analysis: Essential regulators in the heat stress-host interactions. Fish Shellfish Immunol. 2018, 81, 390–398. [Google Scholar] [CrossRef] [PubMed]
- Starega-Roslan, J.; Krol, J.; Koscianska, E.; Kozlowski, P.; Szlachcic, W.J.; Sobczak, K.; Krzyzosiak, W.J. Structural basis of microRNA length variety. Nucleic Acids Res. 2011, 39, 257–268. [Google Scholar] [CrossRef]
- Lee, Y.-Y.; Kim, H.; Kim, V.N. Sequence determinant of small RNA production by DICER. Nature 2023, 615, 323–330. [Google Scholar] [CrossRef]
- Zhong, S.; Ye, X.; Liu, H.; Ma, X.; Chen, X.; Zhao, L.; Huang, G.; Huang, L.; Zhao, Y.; Qiao, Y. MicroRNA sequencing analysis reveals immune responses in hepatopancreas of Fenneropenaeus penicillatus under white spot syndrome virus infection. Fish Shellfish Immunol. 2024, 146, 109432. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Liu, Q.-h.; Yang, B.; Huang, J. Differential expression of microRNAs of Litopenaeus vannamei in response to different virulence WSSV infection. Fish Shellfish Immunol. 2016, 58, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Chen, G.; Hu, J.; Bao, Z.; Wang, M. Transcriptome and microRNAome elucidate the mechanism underlying the immunomodulatory effects of dietary CpG oligodeoxynucleotides (CpG ODNs) in Litopenaeus vannamei. Aquaculture 2024, 593, 741275. [Google Scholar] [CrossRef]
- Zhou, L.; Zhao, F.; Shi, Y.; Liu, H.; Zhang, K.; Zhang, C.; Yin, S.; Ning, X. Comprehensive whole transcriptome analysis reveals specific lncRNA-and circRNA-ceRNA networks of Chinese mitten crab (Eriocheir sinensis) in response to hypoxia stress. Aquaculture 2025, 598, 742017. [Google Scholar] [CrossRef]
- Kanoksinwuttipong, N.; Jaree, P.; Somboonwiwat, K. Shrimp pmo-miR-750 regulates the expression of sarcoplasmic calcium-binding protein facilitating virus infection in Penaeus monodon. Fish Shellfish Immunol. 2022, 129, 74–84. [Google Scholar] [CrossRef]
- Li, Y.; Xu, Q.; Liu, H.; Dai, X. Differential expression of microRNAs in giant freshwater prawn (Macrobrachium rosenbergii) during the infection of Vibrio parahaemolyticus. Fish Shellfish Immunol. 2024, 153, 109827. [Google Scholar] [CrossRef]
- Rollin, M.; Xuereb, B.; Coulaud, R.; Loisel, V.; Poret, A.; Duflot, A.; Le Foll, F.; Picard, C.; Hucher, N. The use of physicochemical properties to assess changes in the cuticle structure of crustaceans: Case of the prawn Palaemon serratus and its moult cycle. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2025, 302, 111801. [Google Scholar] [CrossRef] [PubMed]
- Kornezos, A.; Chia, W. Apical secretion and association of the Drosophila yellow gene product with developing larval cuticle structures during embryogenesis. Mol. Gen. Genet. MGG 1992, 235, 397–405. [Google Scholar] [CrossRef]
- Boyle, R.T.; McNamara, J.C. Association of kinesin and myosin with pigment granules in crustacean chromatophores. Pigment Cell Res. 2006, 19, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Wiriyasermkul, P.; Moriyama, S.; Nagamori, S. Membrane transport proteins in melanosomes: Regulation of ions for pigmentation. Biochim. Biophys. Acta (BBA)-Biomembr. 2020, 1862, 183318. [Google Scholar] [CrossRef]
- Wang, M.-R.; Zhu, X.-J.; Yang, J.-S.; Dai, Z.-M.; Mahmood, K.; Yang, F.; Yang, W.-J. Prawn lipocalin: Characteristics and expressional pattern in subepidermal adipose tissue during reproductive molting cycle. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2007, 147, 222–229. [Google Scholar] [CrossRef] [PubMed]
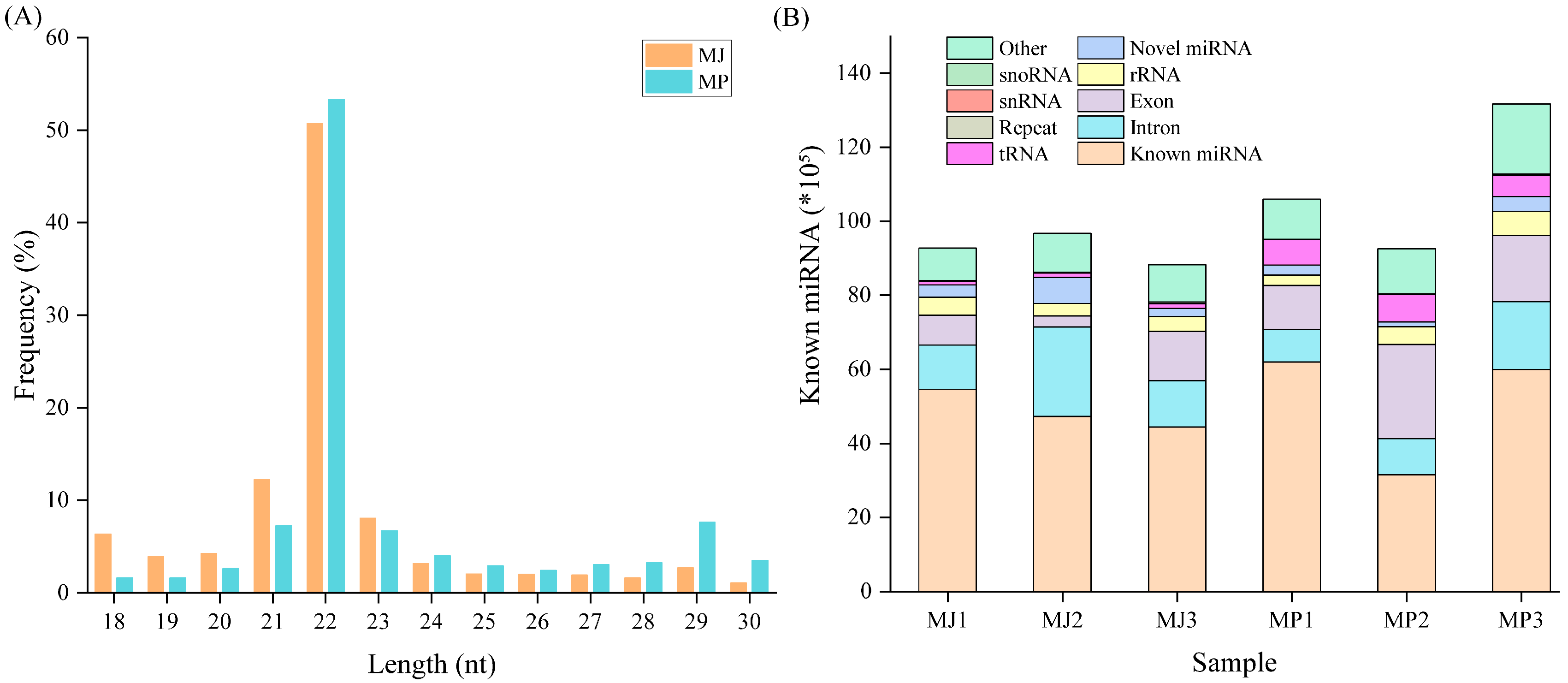



| Sample | MJ1 | MJ2 | MJ3 | MP1 | MP2 | MP3 |
|---|---|---|---|---|---|---|
| Raw reads | 11,762,892 | 11,762,903 | 10,042,741 | 11,728,291 | 10,753,627 | 14,776,981 |
| N% > 10% | 12,242 (0.10%) | 11,449 (0.10%) | 9628 (0.10%) | 11,295 (0.10%) | 10,300 (0.10%) | 11,720 (0.08%) |
| Low quality | 17,612 (0.15%) | 17,041 (0.14%) | 14,243 (0.14%) | 15,837 (0.14%) | 17,496 (0.16%) | 30,422 (0.21%) |
| 3 adapter null or insert null | 35,803 (0.30%) | 365,964 (3.11%) | 302,569 (3.01%) | 193,872 (1.65%) | 166,008 (1.54%) | 199,760 (1.35%) |
| 5 adapter contamine | 31,254 (0.27%) | 15,861 (0.13%) | 26,057 (0.26%) | 35,134 (0.30%) | 28,834 (0.27%) | 58,353 (0.39%) |
| ployA/T/G/C | 6538 (0.06%) | 9144 (0.08%) | 53,716 (0.53%) | 4204 (0.04%) | 9174 (0.09%) | 9638 (0.07%) |
| Clean reads | 11,659,443 (99.12%) | 11,343,444 (96.43%) | 9,636,528 (95.96%) | 11,467,949 (97.78%) | 10,521,815 (97.84%) | 14,467,088 (97.90%) |
| Match | 9,271,883 (78.82%) | 9,674,366 (82.24%) | 8,822,865 (87.85%) | 10,600,620 (90.39%) | 9,254,238 (86.06%) | 13,167,375 (89.11%) |
| Q20 | 99.63% | 99.61% | 99.60% | 99.64% | 99.57% | 99.51% |
| Q30 | 98.64% | 98.52% | 98.54% | 98.68% | 98.38% | 98.21% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, P.; Zhu, J.; Chen, H.; Hu, Q.; Chen, Z.; Li, W.; Yang, T.; Zhu, J.; Yan, B.; Gao, H.; et al. Study on the Regulatory Mechanisms of Carapace Marking Formation in Marsupenaeus japonicus. Animals 2025, 15, 727. https://doi.org/10.3390/ani15050727
Wang P, Zhu J, Chen H, Hu Q, Chen Z, Li W, Yang T, Zhu J, Yan B, Gao H, et al. Study on the Regulatory Mechanisms of Carapace Marking Formation in Marsupenaeus japonicus. Animals. 2025; 15(5):727. https://doi.org/10.3390/ani15050727
Chicago/Turabian StyleWang, Panpan, Jiawei Zhu, Huanyu Chen, Qingyuan Hu, Zhenxiang Chen, Wenjia Li, Ting Yang, Jin Zhu, Binlun Yan, Huan Gao, and et al. 2025. "Study on the Regulatory Mechanisms of Carapace Marking Formation in Marsupenaeus japonicus" Animals 15, no. 5: 727. https://doi.org/10.3390/ani15050727
APA StyleWang, P., Zhu, J., Chen, H., Hu, Q., Chen, Z., Li, W., Yang, T., Zhu, J., Yan, B., Gao, H., & Xing, C. (2025). Study on the Regulatory Mechanisms of Carapace Marking Formation in Marsupenaeus japonicus. Animals, 15(5), 727. https://doi.org/10.3390/ani15050727






