Chloro-Substituted Naphthyridine Derivative and Its Conjugate with Thiazole Orange for Highly Selective Fluorescence Sensing of an Orphan Cytosine in the AP Site-Containing Duplexes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Probe Synthesis
2.3. Fluorescent Measurements
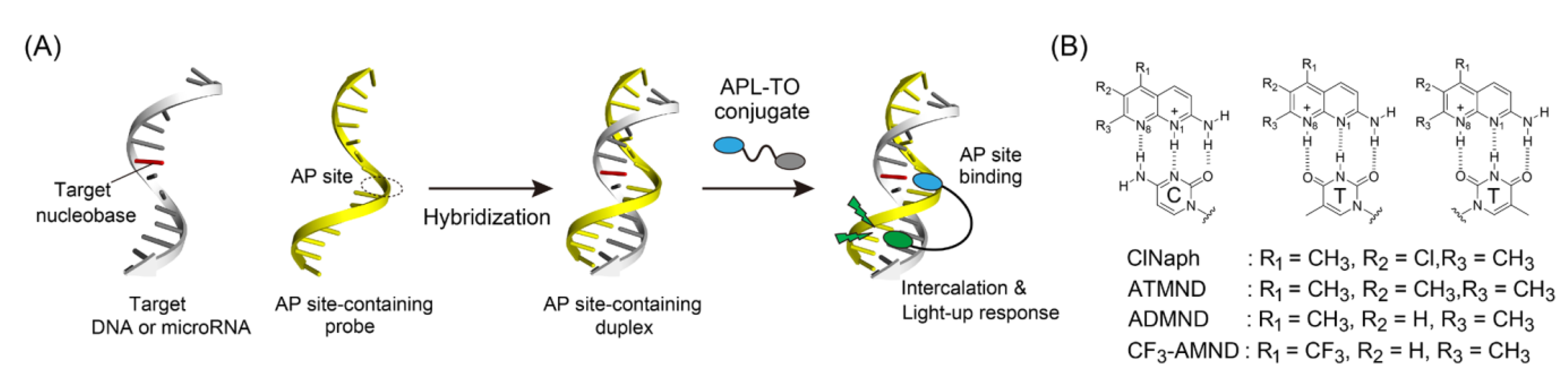
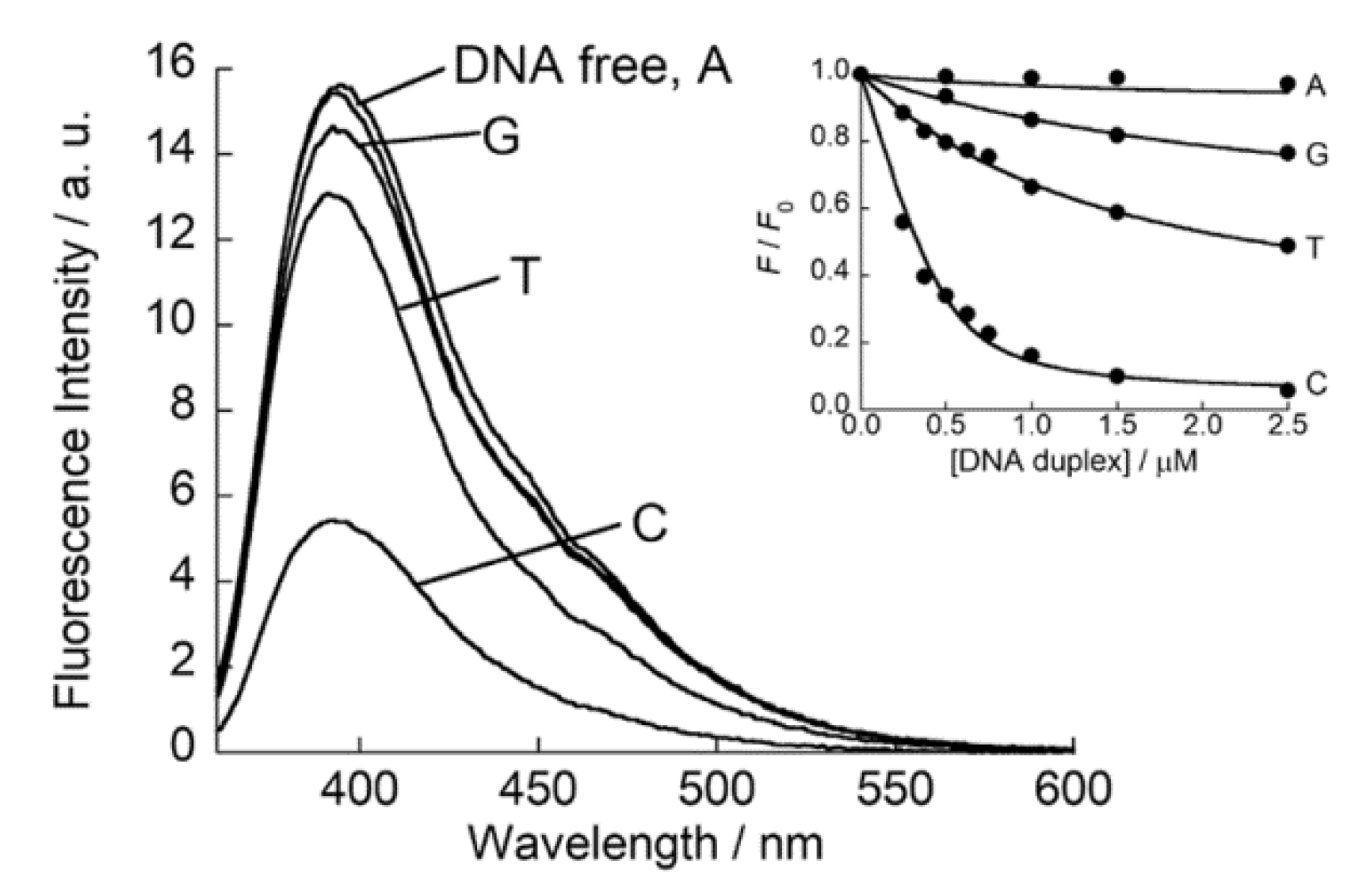
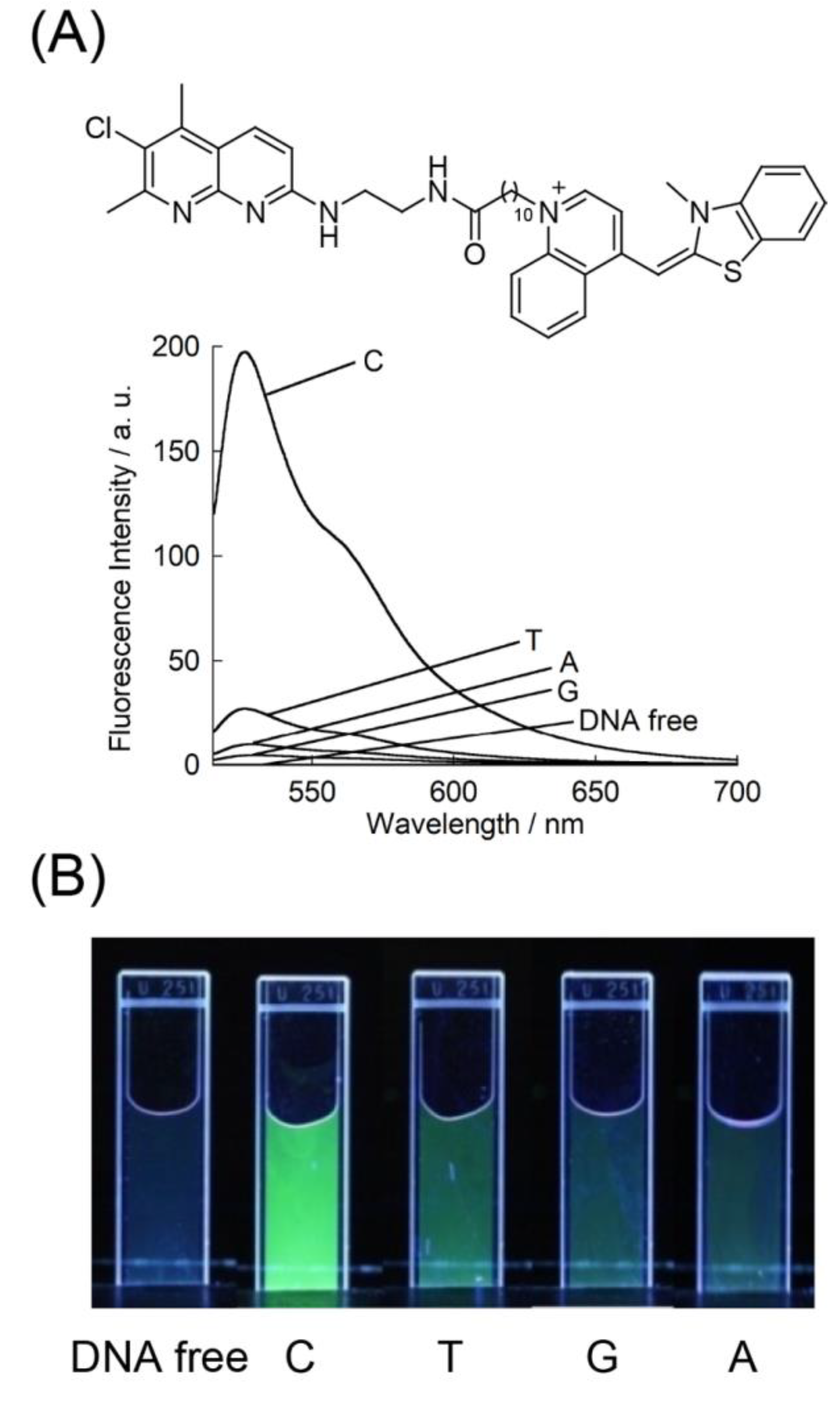
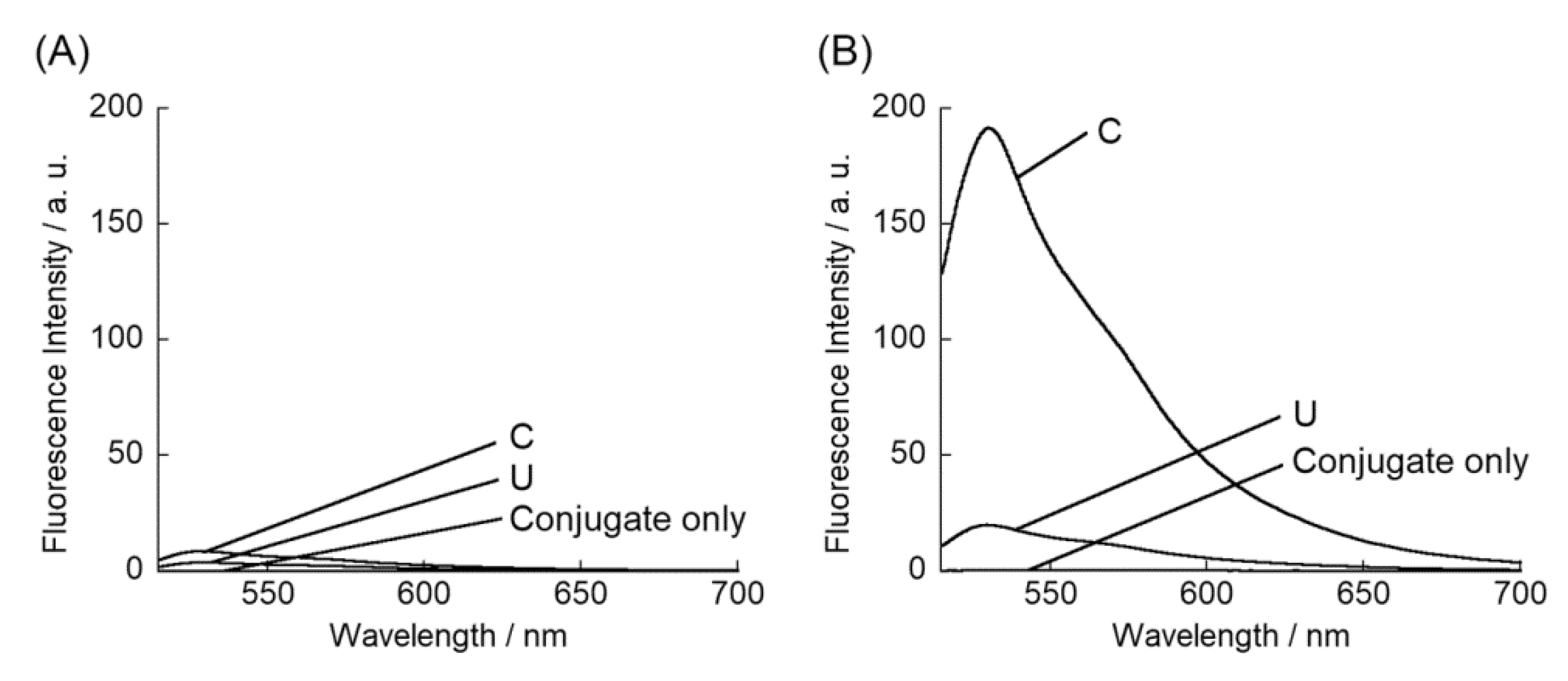
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Granzhan, A.; Kotera, N.; Teulade-Fichou, M.-P. Finding needles in a basestack: Recognition of mismatched base pairs in DNA by small molecules. Chem. Soc. Rev. 2014, 43, 3630–3665. [Google Scholar] [CrossRef] [PubMed]
- Song, G.; Ren, J. Recognition and regulation of unique nucleic acid structures by small molecules. Chem. Commun. 2010, 46, 7283–7294. [Google Scholar] [CrossRef]
- Sato, Y.; Nishizawa, S.; Yoshimoto, K.; Seino, T.; Ichihashi, T.; Morita, K.; Teramae, N. Influence of substituent modifications on the binding of 2-amino-1,8-naphthyridines to cytosine opposite an AP site in DNA duplexes: Thermodynamic characterization. Nucleic Acids Res. 2009, 37, 1411–1422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qing, D.; Xu, C.-Y.; Sato, Y.; Yoshimoto, K.; Nishizawa, S.; Teramae, N. Enhancement of the binding ability of a ligand for nucleobase recognition by introducing a methyl group. Anal. Sci. 2006, 22, 201–203. [Google Scholar]
- Wang, C.-X.; Sato, Y.; Kudo, M.; Nishizawa, S.; Teramae, N. Ratiometric fluorescent signaling of small molecules, environmentally sensitive dye conjugates for detecting single base mutations in DNA. Chem. Eur. J. 2012, 18, 9481–9484. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Ichihashi, T.; Nishizawa, S.; Teramae, N. Strong and selective binding of amiloride to an abasic site in RNA duplexes: Thermodynamic characterization and microRNA detection. Angew. Chem. Int. Ed. 2012, 51, 6339–6372. [Google Scholar] [CrossRef]
- Sato, Y.; Toriyabe, Y.; Nishizawa, S.; Teramae, N. 2,4-Diamino-6,7-dimethylpteridine as a fluorescent ligand for binding andsensing an orphan cytosine in RNA duplexes. Chem. Commun. 2013, 49, 9983–9985. [Google Scholar] [CrossRef]
- Fakhari, M.A.; Rokita, S.E. A new solvatochromic fluorophore for exploring nonpolar environments created by biopolymers. Chem. Commun. 2011, 47, 4222–4224. [Google Scholar] [CrossRef] [PubMed]
- Suda, H.; Kobori, A.; Zhang, J.; Hayashi, G.; Nakatani, K. N′-Bis(3-aminopropyl)-2,7-diamino-1,8-naphthyridine stabilized a single pyrimidine bulge in duplex DNA. Bioorg. Med. Chem. 2005, 13, 4507–4512. [Google Scholar] [CrossRef] [PubMed]
- Verma, R.K.; Takei, F.; Nakatani, K. Synthesis and photophysical properties of fluorescence molecular probe for turn-on-type detection of cytosine bulge DNA. Org. Let. 2016, 18, 3170–3173. [Google Scholar] [CrossRef]
- Arambula, J.F.; Ramisetty, S.R.; Baranger, A.M.; Zimmerman, S.C. A simple ligand that selectively targets CUG trinucleotide repeats and inhibits MBNL protein binding. Proc. Natl. Acad. Sci. USA 2009, 106, 16068–16073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeglis, B.M.; Barton, J.K. A mismatch-selective bifunctional rhodium-oregon green conjugate: A fluorescent probe for mismatched DNA. J. Am. Chem. Soc. 2006, 128, 5654–5655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, T.; Sato, Y.; Iwai, K.; Kuge, S.; Nishizawa, S.; Teramae, N. Synthetic fluorescent probes capable of selective recognition of 3’-overhanging nucleotides for siRNA delivery imaging. Chem. Commun. 2015, 51, 1421–1424. [Google Scholar] [CrossRef]
- Tanabe, T.; Sato, T.; Sato, Y.; Nishizawa, S. Design of a fluorogenic PNA probe capable of simultaneous recognition of 3’-overhang and double-stranded sequences of small interfering RNAs. RSC Adv. 2018, 8, 42095–42099. [Google Scholar] [CrossRef] [Green Version]
- Sato, Y.; Kaneko, M.; Sato, T.; Nakata, S.; Takahashi, Y.; Nishizawa, S. Enhanced binding affinity of siRNA overhang-binding fluorescent probes by conjugation with cationic oligopeptides for improved analysis of the siRNA delivery process. ChemBioChem 2019, 20, 408–414. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Morita, K.; Sato, Y.; Dai, Q.; Nishizawa, S.; Teramae, N. Label-free aptamer-based sensor using abasic site-containing DNA and a nucleobase-specific fluorescent ligand. Chem. Commun. 2009, 42, 6445–6447. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Nishizawa, S.; Teramae, N. Label-free molecular beacon system based on DNAs containing abasic sites and fluorescent ligands that bind abasic sites. Chem. Eur. J. 2011, 17, 11650–11656. [Google Scholar] [CrossRef]
- Sato, Y.; Zhang, Y.; Nishizawa, S.; Seino, T.; Nakamura, K.; Li, M.; Teramae, N. Competitive assay for theophylline based on an abasic site-containing DNA duplex aptamer and a fluorescent ligand. Chem. Eur. J. 2012, 18, 12719–12724. [Google Scholar] [CrossRef]
- Sato, Y.; Kudo, M.; Toriyabe, Y.; Kuchitsu, S.; Wang, C.-X.; Nishizawa, S.; Teramae, N. Abasic site-binding ligands conjugated with cyanine dyes for “off-on” fluorescence sensing of orphan nucleobases in DNA duplexes and DNA-RNA hybrids. Chem. Commun. 2014, 50, 515–517. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Saito, H.; Aoki, D.; Teramae, N.; Nishizawa, S. Lysine linkage in abasic site-binding ligand-thiazole orange conjugates for improved binding affinity to orphan nucleobases in DNA/RNA hybrids. Chem. Commun. 2016, 52, 14446–14449. [Google Scholar] [CrossRef]
- Nygren, J.; Svanvik, N.; Kubista, M. The interactions between the fluorescent dye thiazole orange and DNA. Biopolymers 1998, 46, 39–51. [Google Scholar] [CrossRef]
- Armitage, B.A. Cyanine dye-DNA interactions: Intercalation, groove binding, and aggregation. Top. Curr. Chem. 2005, 253, 55–76. [Google Scholar]
- Dong, H.; Lei, J.; Ding, L.; Wen, Y.; Ju, H.; Zhang, X. MicroRNA: Function, detection, and bioanalysis. Chem. Rev. 2013, 113, 6207–6233. [Google Scholar] [CrossRef]
- Sato, Y.; Zhang, Y.; Seino, T.; Sugimoto, T.; Nishizawa, S.; Teramae, N. Highly selective binding of naphthyridine with a trifluoromethyl group to cytosine opposite an abasic site in DNA duplexes. Org. Biomol. Chem. 2012, 10, 4003–4006. [Google Scholar] [CrossRef] [PubMed]
- Puglisi, J.D.; Tinoco, I. Absorbance melting curves of RNA. Methods Enzymol. 1989, 180, 304–325. [Google Scholar]
- Carreon, J.R.; Stewart, K.M.; Mahon, K.P., Jr.; Shin, S.; Kelly, S.O. Cyanine dye conjugates as probes for live cell imaging. Bioorg. Med. Chem. Lett. 2007, 17, 5182–5185. [Google Scholar] [CrossRef]
- Carreon, J.R.; Mahon, K.P., Jr.; Kelly, S.O. Thiazole orange-peptide conjugates: Sensitivity of DNA binding to chemical structure. Org. Lett. 2004, 6, 517–519. [Google Scholar] [CrossRef]
- Shaw, N.N.; Arya, D.P. Recognition of the unique structure of DNA:RNA hybrids. Biochimie 2008, 90, 1026–1039. [Google Scholar] [CrossRef]
- Johnson, S.M.; Grosshans, H.; Shingara, J.; Byrom, M.; Jarvis, R.; Cheng, A.; Labourier, E.; Reinert, K.L.; Brown, D.; Slack, F.J. RAS is regulated by the let-7 microRNA family. Cell 2005, 120, 635–647. [Google Scholar] [CrossRef] [Green Version]

| Kd/nM | |
|---|---|
| ClNaph | 70 ± 5.3 |
| ADMND b | 164 |
| ATMND b | 53 |
| CF3-AMND c | 1400 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.-x.; Sato, Y.; Sugimoto, T.; Teramae, N.; Nishizawa, S. Chloro-Substituted Naphthyridine Derivative and Its Conjugate with Thiazole Orange for Highly Selective Fluorescence Sensing of an Orphan Cytosine in the AP Site-Containing Duplexes. Appl. Sci. 2020, 10, 4133. https://doi.org/10.3390/app10124133
Wang C-x, Sato Y, Sugimoto T, Teramae N, Nishizawa S. Chloro-Substituted Naphthyridine Derivative and Its Conjugate with Thiazole Orange for Highly Selective Fluorescence Sensing of an Orphan Cytosine in the AP Site-Containing Duplexes. Applied Sciences. 2020; 10(12):4133. https://doi.org/10.3390/app10124133
Chicago/Turabian StyleWang, Chun-xia, Yusuke Sato, Takashi Sugimoto, Norio Teramae, and Seiichi Nishizawa. 2020. "Chloro-Substituted Naphthyridine Derivative and Its Conjugate with Thiazole Orange for Highly Selective Fluorescence Sensing of an Orphan Cytosine in the AP Site-Containing Duplexes" Applied Sciences 10, no. 12: 4133. https://doi.org/10.3390/app10124133





