Establishment of Optogenetic Modulation of cAMP for Analyzing Growth, Biofilm Formation, and Virulence Pathways of Bacteria Using a Light-Gated Cyclase
Abstract
:1. Introduction
2. Materials and Methodology
2.1. Bioinformatics Analysis of LgACs
2.2. Biochemical and Functional Characterization of the Recombinant LgAC
2.3. Measurement of Cyclase Activity of Recombinant LgAC Expressed in E. coli BL21 (λDE3) Cells
2.4. Confocal Microscopy of E. coli Cells Expressing LgAC
2.5. Label-Free Quantitative Proteomics
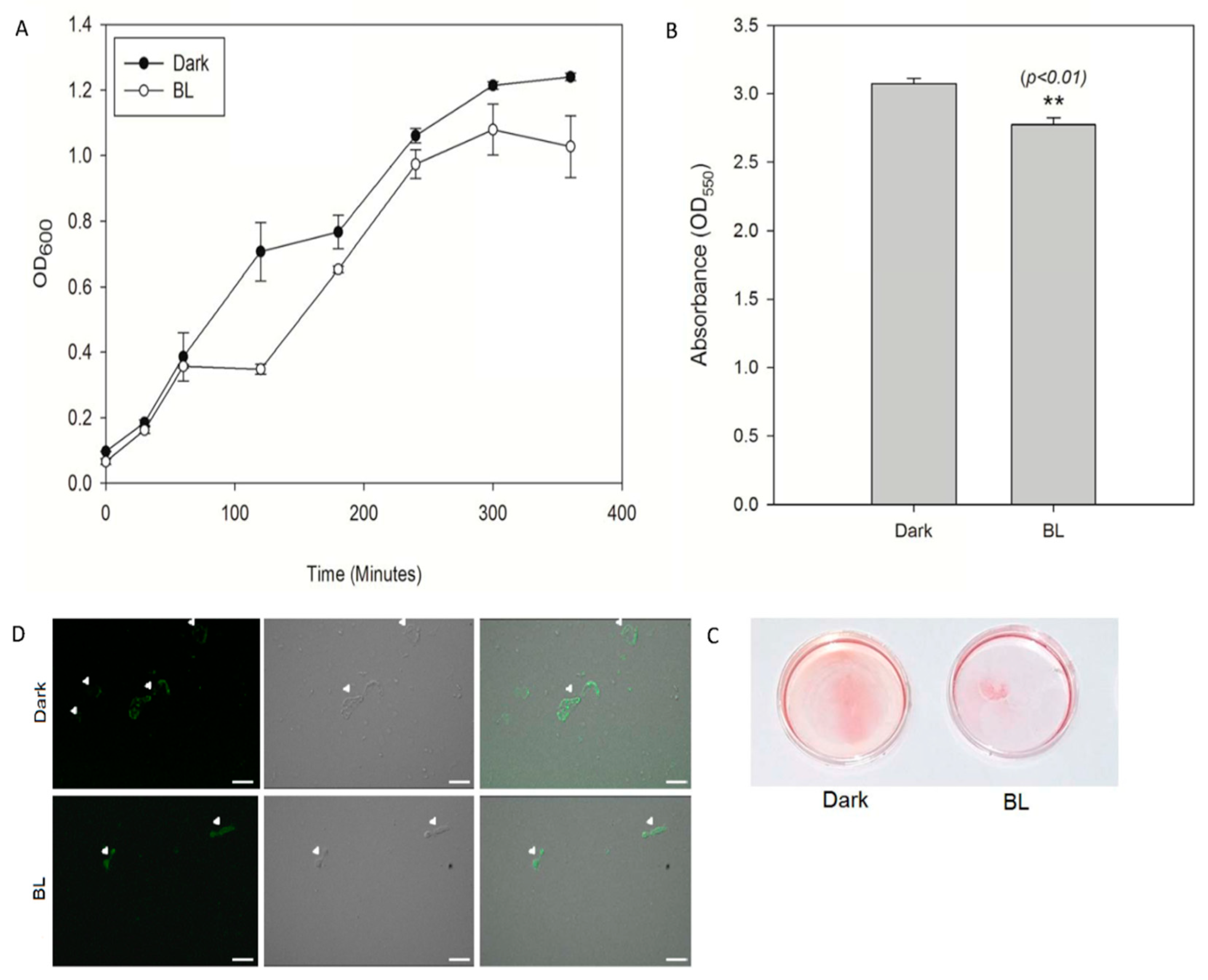
2.6. Growth of E. coli Expressing LgAC
2.7. Biofilm of E. coli Expressing LgAC
3. Results and Discussion
3.1. LgAC is an Optogenetic Modulator of cAMP in E. coli
3.2. Label-Free Quantitative Proteomics Revealed cAMP-Dependent Modulation of Growth, Biofilm Formation, and Virulence in E. coli
3.2.1. LgAC Regulates Growth and Energy Metabolism in E. coli
3.2.2. Functional Expression of LgAC Modulates Biofilm Formation and Associated Processes in E. coli
3.2.3. Optogenetic Modulation of cAMP Regulates Virulence Signaling in E. coli
3.3. Protein Networking Deciphers Functional Links among the DEPs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shimizu, K. Metabolic regulation and coordination of the metabolism in bacteria in response to a variety of growth conditions. Adv. Biochem. Eng. Biotechnol. 2016, 155, 1–54. [Google Scholar]
- You, C.; Okano, H.; Hui, S.; Zhang, Z.; Kim, M.; Gunderson, C.W.; Wang, Y.P.; Lenz, P.; Yan, D.; Hwa, T. Coordination of bacterial proteome with metabolism by cyclic AMP signalling. Nature 2013, 500, 301–306. [Google Scholar] [CrossRef] [Green Version]
- McDonough, K.A.; Rodriguez, A. The myriad roles of cyclic AMP in microbial pathogens: From signal to sword. Nature Rev. Microbiol. 2012, 10, 27–38. [Google Scholar] [CrossRef] [Green Version]
- Görke, B.; Stülke, J. Carbon catabolite repression in bacteria: Many ways to make the most out of nutrients. Nature Rev. Microbiol. 2008, 6, 613–624. [Google Scholar] [CrossRef]
- Hufnagel, D.A.; Evans, M.L.; Greene, S.E.; Pinkner, J.S.; Hultgren, S.J.; Chapman, M.R. The catabolite repressor protein-cyclic AMP complex regulates csgD and biofilm formation in uropathogenic Escherichia coli. J. Bacteriol. 2016, 198, 329–3334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, C.M.; Åberg, A.; Straseviçiene, J.; Emődy, L.; Uhlin, B.E.; Balsalobre, C. Type 1 fimbriae, a colonization factor of uropathogenic Escherichia coli, are controlled by the metabolic sensor CRP-cAMP. PLoS Pathog. 2009, 5, e1000303. [Google Scholar] [CrossRef] [Green Version]
- Sutrina, S.L.; Daniel, K.; Lewis, M.; Charles, N.T.; Anselm, C.K.; Holder, N. Biofilm growth of Escherichia coli is subject to cAMP-dependent and cAMP-independent inhibition. J. Mol. Microbiol. Biotechnol. 2015, 25, 209–225. [Google Scholar] [CrossRef] [PubMed]
- Ritzert, J.T.; Minasov, G.; Embry, R.; Schipma, M.J.; Satchell, K.J. The Cyclic AMP Receptor Protein Regulates Quorum Sensing and Global Gene Expression in Yersinia pestis During Planktonic Growth and Growth in Biofilms. mBio 2019, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alasbahi, R.H.; Melzig, M.F. Forskolin and derivatives as tools for studying the role of cAMP. Pharmazie 2012, 67, 5–13. [Google Scholar] [PubMed]
- Martinez, A.; Gil, C. cAMP-specific phosphodiesterase inhibitors: Promising drugs for inflammatory and neurological diseases. Expert Opin. Ther. Pat. 2014, 24, 1311–1321. [Google Scholar] [CrossRef]
- Levskaya, A.; Weiner, O.D.; Lim, W.A.; Voigt, C.A. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 2009, 461, 997–1001. [Google Scholar] [CrossRef] [Green Version]
- Boyden, E.S.; Zhang, F.; Bamberg, E.; Nagel, G.; Deisseroth, K. Millisecond-time scale, genetically targeted optical control of neural activity. Nature Neurosci. 2005, 8, 1263–1268. [Google Scholar] [CrossRef] [PubMed]
- Tanwar, M.; Khera, L.; Haokip, N.; Kaul, R.; Naorem, A.; Kateriya, S. Modulation of cyclic nucleotide-mediated cellular signaling and gene expression using photoactivated adenylyl cyclase as an optogenetic tool. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Z.; Tanaka, K.F.; Matsunaga, S.; Iseki, M.; Watanabe, M.; Matsuki, N.; Ikegaya, Y.; Koyama, R. Photoactivated adenylyl cyclase (PAC) reveals novel mechanisms underlying cAMP-dependent axonal morphogenesis. Sci. Rep. 2016, 5, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schröder-Lang, S.; Schwärzel, M.; Seifert, R.; Strünker, T.; Kateriya, S.; Looser, J.; Watanabe, M.; Kaupp, U.B.; Hegemann, P.; Nagel, G. Fast manipulation of cellular cAMP level by light in vivo. Nat. Methods 2007, 4, 39–42. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, M.; Sharma, R.; Veetil, S.; Srivastava, S.; Kateriya, S. Modular Diversity of the BLUF Proteins and Their Potential for the Development of Diverse Optogenetic Tools. Appl. Sci. 2019, 9, 3924. [Google Scholar] [CrossRef] [Green Version]
- Stierl, M.; Stumpf, P.; Udwari, D.; Gueta, R.; Hagedorn, R.; Losi, A.; Gärtner, W.; Petereit, L.; Efetova, M.; Schwarzel, M.; et al. Light modulation of cellular cAMP by a small bacterial photoactivated adenylyl cyclase, bPAC, of the soil bacterium Beggiatoa. J. Biol. Chem. 2011, 286, 1181–1188. [Google Scholar] [CrossRef] [Green Version]
- Tanwar, M.; Sharma, K.; Moar, P.; Kateriya, S. Biochemical Characterization of the Engineered Soluble PhotoactivatedGuanylateCyclases from Microbes Expands Optogenetic Tools. Appl. Biochem. Biotechnol. 2018, 185, 1014–1028. [Google Scholar] [CrossRef]
- Geer, L.Y.; Domrachev, M.; Lipman, D.J.; Bryant, S.H. CDART: Protein homology by domain architecture. Genome Res. 2002, 12, 1619–1623. [Google Scholar] [CrossRef] [Green Version]
- Hall, T.A. BIoedit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/ NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- BIOvIA, D.S. Discovery Studio Modeling Environment; Dassault Systemes: San Diego, CA, USA, 2015; Volume 4. [Google Scholar]
- Jhingan, G.D.; Kumari, S.; Jamwal, S.V.; Kalam, H.; Arora, D.; Jain, N.; Kumaar, L.K.; Samal, A.; Rao, K.V.S.; Kumar, D.; et al. Comparative Proteomic Analyses of Avirulent, Virulent, and Clinical Strains of Mycobacterium tuberculosis Identify Strain-specific Patterns. J. Biol. Chem. 2016, 291, 14257–14273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software Environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Oliveros, J.C. VENNY. An Interactive Tool for Comparing Lists with Venn Diagrams. Available online: http://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 18 January 2020).
- Saeed, A.I.; Sharov, V.; White, J.; Li, J.; Liang, W.; Bhagabati, N.; Braisted, J.; Klapa, M.; Currier, T.; Thiagarajan, M.; et al. TM4: A free, open-source system for microarray data management and analysis. Biotechniques 2003, 34, 374–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammer, Ø.; Harper, D.A.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- O’Toole, G.A.; Kolter, R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: A genetic analysis. Mol. Microbiol. 1998, 28, 449–461. [Google Scholar] [CrossRef]
- Christensen, G.D.; Simpson, W.A.; Younger, J.J.; Baddour, L.M.; Barrett, F.F.; Melton, D.M.; Beachey, E.H. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: A quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 1985, 22, 996–1006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kleinboelting, S.; Diaz, A.; Moniot, S.; Van Den Heuvel, J.; Weyand, M.; Levin, L.R.; Buck, J.; Steegborn, C. Crystal structures of human soluble adenylyl cyclase reveal mechanisms of catalysis and of its activation through bicarbonate. Proc. Natl. Acad. Sci. USA 2014, 111, 3727–3732. [Google Scholar] [CrossRef] [Green Version]
- Kolb, A.; Busby, S.; Buc, H.; Garges, S.; Adhya, S. Transcriptional regulation by cAMP and its receptor protein. Ann. Rev. Biochem. 1993, 62, 749–797. [Google Scholar] [CrossRef]
- Bettenbrock, K.; Sauter, T.; Jahreis, K.; Kremling, A.; Lengeler, J.W.; Gilles, E.D. Correlation between growth rates, EIIACrr phosphorylation, and intracellular cyclic AMP levels in Escherichia coli K-12. J. Bacteriol. 2007, 189, 6891–6900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.H.; Shen, R.; Olcott, M.C.; Rajagopal, I.; Mathews, C.K. Adenylate kinase of Escherichia coli, a component of the phage T4 dNTPsynthetase complex. J. Biol. Chem. 2005, 280, 28221–28229. [Google Scholar] [CrossRef] [Green Version]
- Thach, T.T.; Luong, T.T.; Lee, S.; Rhee, D.K. Adenylate kinase from Streptococcus pneumoniae is essential for growth through its catalytic activity. FEBS Open Bio 2014, 4, 672–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fox, D.K.; Meadow, N.D.; Roseman, S. Phosphate transfer between acetate kinase and Enzyme I of the bacterial phosphotransferase system. J. Biol. Chem. 1986, 261, 13498–13503. [Google Scholar] [PubMed]
- Tiwari, S.; Barh, D.; Imchen, M.; Rao, E.; Kumavath, R.K.; Seenivasan, S.P.; Jaiswal, A.K.; Jamal, S.B.; Kumar, V.; Ghosh, P.; et al. Acetate Kinase (AcK) is Essential for Microbial Growth and Betel-derived Compounds Potentially Target AcK, PhoP and MDR Proteins in M. tuberculosis, V. cholerae and Pathogenic, E. coli: An in silico and in vitro Study. Curr. Top. Med. Chem. 2018, 18, 2731. [Google Scholar] [CrossRef]
- Yan, D. Protection of the glutamate pool concentration in enteric bacteria. Proc. Natl. Acad. Sci. USA 2007, 104, 9475–9480. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, T.P.; Shauger, A.E.; Kustu, S. Salmonella typhimurium apparently perceives external nitrogen limitation as internal glutamine limitation. J. Mol. Biol. 1996, 259, 589–607. [Google Scholar] [CrossRef] [PubMed]
- Aldarini, N.; Alhasawi, A.A.; Thomas, S.C.; Appanna, V.D. The role of glutamine synthetase in energy production and glutamine metabolism during oxidative stress. Antonie van Leeuwenhoek. Int. J. Gen. Mol. Microbiol. 2017, 110, 629–639. [Google Scholar]
- Auger, C.; Appanna, V.D. A novel ATP-generating machinery to counter nitrosative stress is mediated by substrate-level phosphorylation. Biochim. Biophys. Acta 2015, 1850, 43–50. [Google Scholar] [CrossRef]
- Ashraf, K.U.; Josts, I.; Mosbahi, K.; Kelly, S.M.; Byron, O.; Smith, B.O.; Walker, D. The Potassium Binding Protein Kbp Is a Cytoplasmic Potassium Sensor. Structure 2016, 24, 741–749. [Google Scholar] [CrossRef] [Green Version]
- Woolstenhulme, C.J.; Guydosh, N.R.; Green, R.; Buskirk, A.R. High-precision analysis of translational pausing by ribosome profiling in bacteria lacking EFP. Cell Rep. 2015, 11, 13–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johansson, M.; Ieong, K.W.; Trobro, S.; Strazewski, P.; Åqvist, J.; Pavlov, M.Y.; Ehrenberg, M. pH-sensitivity of the ribosomal peptidyl transfer reaction dependent on the identity of the A-site aminoacyl-tRNA. Proc. Natl. Acad. Sci. USA 2011, 108, 79–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doerfel, L.K.; Rodnina, M.V. Elongation factor P: Function and effects on bacterial fitness. Biopolymers 2013, 99, 837–845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuo, J.T.; Chang, Y.J.; Tseng, C.P. Growth rate regulation of lac operon expression in Escherichia coli is cyclic AMP dependent. FEBS Lett. 2003, 553, 397–402. [Google Scholar] [CrossRef] [Green Version]
- Diaz, M.R.; King, J.M.; Yahr, T.L. Intrinsic and extrinsic regulation of type III secretion gene expression in Pseudomonas aeruginosa. Front. Microbiol. 2011, 2, 89. [Google Scholar] [CrossRef] [Green Version]
- Fong, J.C.; Yildiz, F.H. Interplay between cyclic AMP-cyclic AMP receptor protein and cyclic di-GMP signaling in Vibrio cholerae biofilm formation. J. Bacteriol. 2008, 190, 6646–6659. [Google Scholar] [CrossRef] [Green Version]
- Liang, W.; Pascual-Montano, A.; Silva, A.J.; Benitez, J.A. The cyclic AMP receptor protein modulates quorum sensing, motility and multiple genes that affect intestinal colonization in Vibrio cholerae. Appl. Environ. Microbiol. 2007, 153, 2964–2975. [Google Scholar] [CrossRef] [Green Version]
- Laverty, G.; Gorman, S.P.; Gilmore, B.F. Biomolecular mechanisms of Pseudomonas aeruginosa and Escherichia coli biofilm formation. Pathogens 2014, 3, 596–632. [Google Scholar] [CrossRef] [Green Version]
- Amores, G.R.; De Las Heras, A.; Sanches-Medeiros, A.; Elfick, A.; Silva-Rocha, R. Systematic identification of novel regulatory interactions controlling biofilm formation in the bacterium Escherichia coli. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Guttenplan, S.B.; Shaw, S.; Kearns, D.B. The cell biology of peritrichous flagella in Bacillus subtilis. Mol. Microbiol. 2013, 87, 211–229. [Google Scholar] [CrossRef] [Green Version]
- Pratt, L.A.; Kolter, R. Genetic analysis of Escherichia coli biofilm formation: Roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 1998, 30, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Suriyanarayanan, T.; Periasamy, S.; Lin, M.H.; Ishihama, Y.; Swarup, S. FlagellinFliC phosphorylation affects type 2 protease secretion and biofilm dispersal in Pseudomonas aeruginosa PAO1. PLoS ONE 2016, 11, 1–19. [Google Scholar] [CrossRef]
- Ibanez-Ruiz, M.; Robbe-Saule, V.; Hermant, D.; Labrude, S.; Norel, F. Identification of RpoS(S)-Regulated Genes in Salmonella entericaSerovarTyphimurium. J. Bacteriol. 2000, 182, 5749–5756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schembri, M.A.; Kjærgaard, K.; Klemm, P. Global gene expression in Escherichia coli biofilms. Mol. Microbiol. 2003, 48, 253–267. [Google Scholar] [CrossRef] [PubMed]
- An, S.Q.; Caly, D.L.; McCarthy, Y.; Murdoch, S.L.; Ward, J.; Febrer, M.; Dow, J.M.; Ryan, R.P. Novel Cyclic di-GMP Effectors of the YajQ Protein Family Control Bacterial Virulence. PLoS Pathog. 2014, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chai, Y.; Chu, F.; Kolter, R.; Losick, R. Bistability and biofilm formation in Bacillus subtilis. Mol. Microbiol. 2008, 67, 254–263. [Google Scholar] [CrossRef] [Green Version]
- Yepes, A.; Schneider, J.; Mielich, B.; Koch, G.; García-Betancur, J.C.; Ramamurthi, K.S.; Vlamakis, H.; López, D. The biofilm formation defect of a Bacillus subtilisflotillin-defective mutant involves the protease FtsH. Mol. Microbiol. 2012, 86, 457–471. [Google Scholar] [CrossRef] [Green Version]
- Kalia, D.; Merey, G.; Nakayama, S.; Zheng, Y.; Zhou, J.; Luo, Y.; Guo, M.; Roembke, B.T.; Sintim, H.O. Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP,(p) ppGpp signaling in bacteria and implications in pathogenesis. Chem. Soc. Rev. 2013, 42, 305–341. [Google Scholar] [CrossRef]
- McCue, L.A.; McDonough, K.A.; Lawrence, C.E. Functional classification of cNMP-binding proteins and nucleotide cyclases with implications for novel regulatory pathways in Mycobacterium tuberculosis. Genome Res. 2000, 10, 204–219. [Google Scholar] [CrossRef] [Green Version]
- Bhandari, V.; Wong, K.S.; Zhou, J.L.; Mabanglo, M.F.; Batey, R.A.; Houry, W.A. The Role of ClpP Protease in Bacterial Pathogenesis and Human Diseases. ACS Chem. Biol. 2018, 13, 1413–1425. [Google Scholar] [CrossRef]
- Yamamoto, T.; Sashinami, H.; Takaya, A.; Tomoyasu, T.; Matsui, H.; Kikuchi, Y.; Hanawa, T.; Kamiya, S.; Nakane, A. Disruption of the genes for ClpXP protease in Salmonella enterica serovarTyphimurium results in persistent infection in mice, and development of persistence requires endogenous gamma interferon and tumor necrosis factor alpha. Infect. Immun. 2001, 69, 3164–3174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knudsen, G.M.; Olsen, J.E.; Aabo, S.; Barrow, P.; Rychlik, I.; Thomsen, L.E. ClpP deletion causes attenuation of Salmonella typhimurium virulence through mis-regulation of rpoS and indirect control of CsrA and the SPI genes. Microbiology (UK) 2013, 159, 1497–1509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, H.; Li, N.; Dong, Z.; Surette, M.G.; Duan, K. The YebC family protein PA0964 negatively regulates the Pseudomonas aeruginosa quinolone signal system and pyocyanin production. J. Bacteriol. 2008, 190, 6217–6227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Passos da Silva, D.; Schofield, M.C.; Parsek, M.R.; Tseng, B.S. An update on the sociomicrobiology of quorum sensing in gram-negative biofilm development. Pathogens 2017, 6, 51. [Google Scholar] [CrossRef] [PubMed]
- Lopez, J.; Feldman, M.F. Expanding the molecular weaponry of bacterial species. J. Biol. Chem. 2018, 293, 1515–1516. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, V.S.; Douzi, B.; Durand, E.; Roussel, A.; Cascales, E.; Cambillau, C. Towards a complete structural deciphering of Type VI secretion system. Curr. Opin. Struct. Biol. 2018, 49, 77–84. [Google Scholar] [CrossRef] [Green Version]
- Liang, W.; Sultan, S.Z.; Silva, A.J.; Benitez, J.A. Cyclic AMP post-transcriptionally regulates the biosynthesis of a major bacterial autoinducer to modulate the cell density required to activate quorum sensing. FEBS Lett. 2008, 582, 3744–3750. [Google Scholar] [CrossRef] [Green Version]
- Neumann, W.; Hadley, R.C.; Nolan, E.M. Transition metals at the host–pathogen interface: How Neisseria exploit human metalloproteins for acquiring iron and zinc. Essays Biochem. 2017, 61, 211–223. [Google Scholar]
- Arenas, F.A.; Díaz, W.A.; Leal, C.A.; Pérez-Donoso, J.M.; Imlay, J.A.; Vásquez, C.C. The Escherichia coli btuE gene, encodes a glutathione peroxidase that is induced under oxidative stress conditions. Biochem. Biophys. Res. Commun. 2010, 398, 690–694. [Google Scholar] [CrossRef] [Green Version]
- Tondo, M.L.; Petrocelli, S.; Ottado, J.; Orellano, E.G. The monofunctional catalase kate of Xanthomonasaxonopodispv. citri is required for full virulence in citrus plants. PLoS ONE 2010, 5, e10803. [Google Scholar] [CrossRef] [Green Version]
- Subramanian Vignesh, K.; Deepe, G.S. Immunological orchestration of zinc homeostasis: The battle between host mechanisms and pathogen defenses. Arch. Biochem. Biophys. 2016, 611, 66–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capdevila, D.A.; Wang, J.; Giedroc, D.P. Bacterial strategies to maintain zinc metallostasis at the host-pathogen interface. J. Biol. Chem. 2016, 291, 20858–20868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michel, A.; Agerer, F.; Hauck, C.R.; Herrmann, M.; Ullrich, J.; Hacker, J.; Ohlsen, K. Global regulatory impact of ClpP protease of Staphylococcus aureus on regulons involved in virulence, oxidative stress response, autolysis, and DNA repair. J. Bacteriol. 2006, 188, 5783–5796. [Google Scholar] [CrossRef] [PubMed] [Green Version]

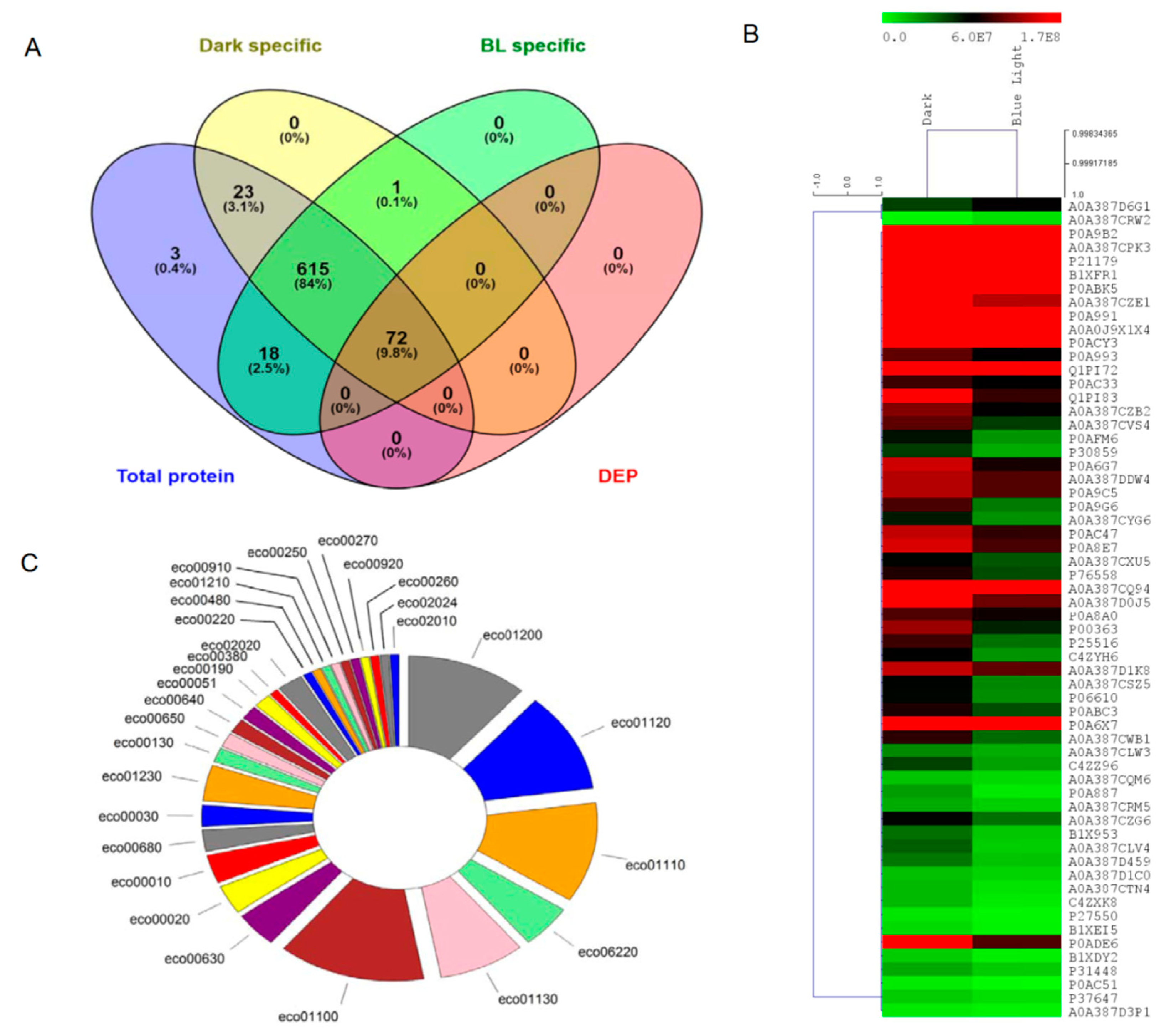

| Accession No. | Differentially Expressed Protein (DEPs) | Abv. | Log Student’s t-Test p-Value | Abundance (Log10) | |
|---|---|---|---|---|---|
| Dark Blue Light | |||||
| P0A9B2 | Glyceraldehyde-3-phosphate dehydrogenase A | GapA | 2.51 | 9.62 | 9.43 |
| A0A387CPK3* | Phosphoenolpyruvate-protein phosphotransferase | PTS* | 2.22 | 8.45 | 8.23 |
| P21179 | Catalase | KatE$ | 2.03 | 9.06 | 8.84 |
| B1XFR1* | Adenylate kinase | Adk* | 2.39 | 8.50 | 8.28 |
| P0ABK5 | Cysteine synthase A | CysK | 2.29 | 8.54 | 8.41 |
| A0A387CZE1* | Acetate kinase | AckA* | 2.01 | 8.32 | 8.11 |
| P0A991 | Fructose-bisphosphate aldolase class 1 | FbaB | 1.98 | 8.81 | 8.68 |
| P0ACY3 | Uncharacterized protein | YeaG# | 1.89 | 8.54 | 8.22 |
| P0A993 | Fructose-1,6-bisphosphatase class 1 | Fbp | 1.96 | 7.99 | 7.79 |
| Q1PI72 | Isocitrate dehydrogenase (Fragment) | Icd | 1.87 | 8.54 | 8.32 |
| P0AC33 | Fumarate hydratase class I, aerobic | FumA | 1.90 | 7.93 | 7.80 |
| A0A387CZB2 | 3-oxoacyl-ACP synthase I | KasA | 2.15 | 8.04 | 7.80 |
| A0A387CVS4 | Citrate synthase | GltA | 2.39 | 7.98 | 7.65 |
| P0AFM6 | Phage shock protein A | PspA | 2.57 | 7.73 | 7.40 |
| P30859 | Putative ABC transporter arginine-binding protein 2 | ArtI | 1.97 | 7.66 | 7.30 |
| P0A6G7 | ATP-dependent Clp protease proteolytic subunit | ClpP$ | 2.66 | 8.14 | 7.86 |
| P0A9C5 | Glutamine synthetase | GlnA* | 2.10 | 8.11 | 7.96 |
| P0A9G6 | Isocitrate lyase | AceA | 2.05 | 7.95 | 7.50 |
| A0A387CYG6 | Pyruvate dehydrogenase [ubiquinone] | Pdh | 2.28 | 7.72 | 7.42 |
| P0AC47 | Fumarate reductase iron-sulfur subunit | FrdB | 2.02 | 8.12 | 7.91 |
| P0A8E7 | UPF0234 protein | YajQ# | 1.89 | 8.15 | 7.94 |
| A0A387CXU5 | Glutamine--tRNA ligase | GlnS | 2.14 | 7.77 | 7.59 |
| P76558 | NADP-dependent malic enzyme | MaeB | 2.75 | 7.88 | 7.62 |
| A0A387CQ94 | Oxidative stress defense protein | nk$ | 2.06 | 8.64 | 8.48 |
| A0A387D0J5 | Flagellin | FliC# | 2.39 | 8.30 | 8.00 |
| P0A8A0 | Probable transcriptional regulatory protein | YebC$ | 2.33 | 7.96 | 7.84 |
| P00363 | Fumarate reductase flavoprotein subunit | FrdA | 3.07 | 8.07 | 7.70 |
| P25516 | Aconitate hydratase A | AcnA | 2.27 | 7.93 | 7.52 |
| C4ZYH6 | Phenylalanine--tRNA ligase alpha subunit | PheS | 2.14 | 7.80 | 7.41 |
| A0A387D1K8 | Transaldolase | Tal | 2.51 | 8.12 | 7.98 |
| A0A387CSZ5 | Transketolase | Tkt | 2.66 | 7.77 | 7.45 |
| P06610 | Thioredoxin/glutathione peroxidase | BtuE$ | 1.88 | 7.76 | 7.43 |
| P0ABC3 | Modulator of FtsH protease | HflC# | 2.82 | 7.87 | 7.61 |
| P0A6X7 | Integration host factor subunit alpha | IhfA# | 3.01 | 8.61 | 8.43 |
| A0A387D6G1 | ATP-dependent protease subunit | HslV | 2.53 | 7.65 | 7.80 |
| A0A387CWB1 | Aldehyde reductase | Ahr | 2.59 | 7.90 | 7.55 |
| A0A387CLW3 | Glycogen synthase | GlgA | 2.03 | 7.45 | 7.29 |
| C4ZZ96 | NH (3)-dependent NAD(+) synthetase | NadE | 2.62 | 7.65 | 7.35 |
| A0A387CQM6 | Tryptophan--tRNA ligase | TrpS | 1.89 | 7.15 | 6.95 |
| P0A887 | Ubiquinone/menaquinone biosynthesis C-methyltransferase | UbiE | 3.37 | 7.36 | 6.74 |
| A0A387CRM5 | AsmA family protein | AsmA | 1.95 | 7.30 | 7.05 |
| A0A387CZG6 | 1,4-dihydroxy-2-naphthoyl-CoA synthase | MenB | 1.91 | 7.76 | 7.52 |
| B1X953 | ADP-L-glycero-D-manno-heptose-6-epimerase | HldD | 2.02 | 7.53 | 7.12 |
| A0A387CLV4 | Oxidoreductase | Nk | 1.99 | 7.58 | 7.06 |
| A0A387D459 | D-alanyl-D-alanine carboxypeptidase | Nk | 2.00 | 7.50 | 7.15 |
| A0A387D1C0 | Dihydroxy-acid dehydratase | IlvD | 1.95 | 7.18 | 7.02 |
| A0A387CTN4 | Elongation factor P-like protein | Efp* | 2.82 | 7.22 | 6.85 |
| C4ZXK8 | L-threonine 3-dehydrogenase | Tdh | 2.29 | 7.22 | 6.71 |
| P27550 | Acetyl-coenzyme A synthetase | Acs | 2.90 | 6.80 | 6.31 |
| B1XEI5 | tRNA-modifying protein | YgfZ | 3.06 | 6.93 | 6.47 |
| P0ADE6 | Potassium binding protein | Kbp* | 2.08 | 8.41 | 7.96 |
| B1XDY2 | Esterase | FrsA | 2.06 | 7.09 | 6.61 |
| P31448 | Uncharacterized symporter | YidK | 2.84 | 7.31 | 7.09 |
| P0AC51 | Zinc uptake regulation protein | Zur$ | 2.48 | 6.68 | 6.13 |
| P37647 | 2-dehydro-3-deoxygluconokinase | KdgK | 2.13 | 7.09 | 6.88 |
| A0A387CRW2 | Lipoprotein | Nk | 1.89 | 6.24 | 6.87 |
| A0A387D3P1 | Endonuclease V | Nfi | 3.40 | 6.74 | 6.51 |
| KEGG TERM | KEGG ID | Input Number | p-Value |
|---|---|---|---|
| Carbon metabolism | eco01200 | 14 | 1.76 × 10−13 |
| Microbial metabolism in diverse environments | eco01120 | 14 | 4.91 × 10−9 |
| Biosynthesis of secondary metabolites | eco01110 | 14 | 6.47 × 10−8 |
| Pyruvate metabolism | eco00620 | 6 | 5.29 × 10−6 |
| Biosynthesis of antibiotics | eco01130 | 10 | 5.58 × 10−6 |
| Metabolic pathways | eco01100 | 17 | 1.61 × 10−5 |
| Glyoxylate and dicarboxylate metabolism | eco00630 | 5 | 2.99 × 10−5 |
| Citrate cycle (TCA cycle) | eco00020 | 4 | 0.000102 |
| Glycolysis / Gluconeogenesis | eco00010 | 4 | 0.000487 |
| Methane metabolism | eco00680 | 3 | 0.001768 |
| Pentose phosphate pathway | eco00030 | 3 | 0.002336 |
| Biosynthesis of amino acids | eco01230 | 5 | 0.002993 |
| Ubiquinone and other terpenoid-quinone biosynthesis | eco00130 | 2 | 0.013632 |
| Butanoate metabolism | eco00650 | 2 | 0.040092 |
| Propanoate metabolism | eco00640 | 2 | 0.041988 |
| Fructose and mannose metabolism | eco00051 | 2 | 0.047874 |
| Oxidative phosphorylation | eco00190 | 2 | 0.047874 |
| Tryptophan metabolism | eco00380 | 1 | 0.079139 |
| Two-component system | eco02020 | 3 | 0.127058 |
| Arginine biosynthesis | eco00220 | 1 | 0.145133 |
| Glutathione metabolism | eco00480 | 1 | 0.152176 |
| 2-Oxocarboxylic acid metabolism | eco01210 | 1 | 0.199917 |
| Nitrogen metabolism | eco00910 | 1 | 0.199917 |
| Alanine, aspartate and glutamate metabolism | eco00250 | 1 | 0.232407 |
| Cysteine and methionine metabolism | eco00270 | 1 | 0.238749 |
| Sulfur metabolism | eco00920 | 1 | 0.245041 |
| Glycine, serine and threonine metabolism | eco00260 | 1 | 0.263613 |
| Quorum sensing | eco02024 | 1 | 0.386875 |
| ABC transporters | eco02010 | 1 | 0.766284 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaushik, M.S.; Pati, S.R.; Soni, S.; Mishra, A.; Sushmita, K.; Kateriya, S. Establishment of Optogenetic Modulation of cAMP for Analyzing Growth, Biofilm Formation, and Virulence Pathways of Bacteria Using a Light-Gated Cyclase. Appl. Sci. 2020, 10, 5535. https://doi.org/10.3390/app10165535
Kaushik MS, Pati SR, Soni S, Mishra A, Sushmita K, Kateriya S. Establishment of Optogenetic Modulation of cAMP for Analyzing Growth, Biofilm Formation, and Virulence Pathways of Bacteria Using a Light-Gated Cyclase. Applied Sciences. 2020; 10(16):5535. https://doi.org/10.3390/app10165535
Chicago/Turabian StyleKaushik, Manish Singh, Swaroop Ranjan Pati, Shivanika Soni, Ayushi Mishra, Kumari Sushmita, and Suneel Kateriya. 2020. "Establishment of Optogenetic Modulation of cAMP for Analyzing Growth, Biofilm Formation, and Virulence Pathways of Bacteria Using a Light-Gated Cyclase" Applied Sciences 10, no. 16: 5535. https://doi.org/10.3390/app10165535
APA StyleKaushik, M. S., Pati, S. R., Soni, S., Mishra, A., Sushmita, K., & Kateriya, S. (2020). Establishment of Optogenetic Modulation of cAMP for Analyzing Growth, Biofilm Formation, and Virulence Pathways of Bacteria Using a Light-Gated Cyclase. Applied Sciences, 10(16), 5535. https://doi.org/10.3390/app10165535






