1. Introduction
During the last few years, Deep Learning (DL) and especially Convolutional Neural Networks (CNNs) have revolutionized computer vision and set new standards for various challenging tasks, such as image classification and semantic segmentation. Since these tasks are also shared in diagnostics, pathology, high-throughput screening, cellular and molecular image analysing and more, the thriving of deep learning was also witnessed in the field of biomedical image analysis [
1,
2].
However, compared to 2D images mostly used in computer vision, image data encountered in the biomedical field are often volumetric. The substantial difficulties in annotating and interpreting of these 3D volumetric data generally result in a much smaller training set than that of computer vision tasks. In addition, in order to explore the 3D context information effectively, which is essential in volumetric data analysing, much effort has to be made in the designing of the network. The current efforts often lead to either a significant increase in the amount of learnable parameters or the complexity in the network design and training. When dealing with large 3D image volumes, the computational cost, as well as the memory requirement will also become damaging even with cutting-edge computational hardware. Therefore, how to explore the 3D contextual information effectively and train an efficient volumetric network with limited training data are still open problems in the biomedical image computing community.
In order to process 3D volumes using CNNs, many schemes have been proposed in the past few years. One straightforward solution is to apply the conventional 2D CNNs on each volume slice separately [
3]. Apparently, this method is a non-optimal use of the volumetric data since the contextual information along the third dimension is disregarded. To make a better use of the 3D context, the tri-planer schemes [
4] suggested applying 2D CNNs on three orthogonal planes (i.e., xy, xz and yz planes). Since the inter-slice information is utilized through a selective choosing of input data, only a small fraction of 3D information is explored [
5]. By viewing the adjacent volume slices as a time series, the Recurrent Neural Network (RNN) was adopted to distil the 3D context from a sequence of abstracted 2D context [
6]. Due to the asymmetric nature of network design, the intra- and inter-slice information cannot be treated and explored equally.
Currently, the 3D CNNs that take 3D convolution kernels as the basic unit [
7,
8] and their hybrid with 2D CNNs [
9] have become the most popular choices in volumetric networks’ design. In addition to impressive 3D context mining ability, the popularity of 3D CNNs is also due to the simple structure nature of 3D operations (e.g., 3D convolutions, 3D pooling and 3D up-convolutions) and their similar usage to corresponding 2D operations [
2,
8]. As a commonly adopted strategy, a 3D CNN can be constructed from the modern 2D CNNs by replacing the 2D operations with their 3D counterparts.
However, the utilization of 3D operations, especially the 3D convolutional kernels, will introduce a huge increase in the amount of trainable parameters, as well as significant memory and computational requirements [
10]. Considering the limited training data often encountered in biomedical tasks, to avoid overfitting, a trade-off has to be made between the scale of the network and its representational power. With these limitations, the existing 3D CNNs tend to contain much fewer layers than modern 2D CNNs in computer vision tasks. For example, 3D U-net has 10 convolutional layers in the encoder part, whereas ResNet [
11] usually has 101 convolutional layers. Since the impact of the network’s depth has been extensively demonstrated to produce improved results in computer vision [
11,
12], there was still much room to explore the potential of 3D CNNs and improve their representational power.
In this paper, instead of modifying the network’s overall architecture to circumvent the trade-off between model size and its representational power, we address this dilemma by looking into the very basic unit of 3D CNNs: 3D convolutional kernels, and propose replacing them with a compact module that possesses better parameter efficiency and stronger nonlinear representational power. We named the proposed module 3D Dense Separated Convolution (3D-DSC), and
Figure 1 illustrates its layout schematically.
The 3D-DSC module was constructed by a series of densely connected 1D filters. Decomposing 3D kernels into 1D filters alleviated the risk of overfitting by removing the redundancy within 3D kernels in a topologically constrained manner, while providing the infrastructure for deepening the network. The nonlinear layers inserted between 1D filters were responsible for the boosting of the block’s nonlinearity, as well as its representational power. The dense connections between 1D filters ensured efficient propagation of information and gradient flow, thus facilitating the training of deepened network. Finally, the 1 × 1 × 1 convolution attached at the end of the block acted as a bottleneck layer to reduce the number of output feature volumes. Compared with direct 3D convolutions, the introduction of 3D-DSC not only effectively deepened the network, thus improving its representational power, but also considerably reduced the number of learnable parameters. This feature is especially useful when training data are limited. In addition, since 3D-DSC did not change the number of input and output feature maps of the convolutional layers, it could be directly used to replace 3D convolutions to boost the network’s performance without modifying its overall architecture.
We evaluated the effectiveness and efficiency of 3D-DSC on volumetric image classification and segmentation, which are two challenging tasks often encountered in biomedical image computing. The results on both tasks showed that significant performance improvements could be consistently obtained with comparable or even fewer parameters than the original 3D convolution version.
Our main contributions are summarized as follows.
(1) We propose an effective strategy for alleviating the overfitting problem while enabling the effective training of a much deeper network in volumetric image analysis, especially for the cases with limited training samples. It is demonstrated that a significant accuracy improvement can be achieved in both classification and segmentation tasks with a similar number of parameters.
(2) The dense connections are introduced between 1D filters in our work to facilitate the training of the deeper network, while it is impractical to introduce dense connections for paralleled 2D and 1D filters. In addition, with nonlinear layers inserted, the effective depth of the network, as well as its representational power can be considerably increased without increasing the number of parameters.
(3) The proposed 3D-DSC is not limited to any specific architecture or application, and it can be used to boost the performance by directly substituting the original 3D convolutional kernels.
2. Related Work
Maximizing the potential of training data is a major goal of supervised machine learning. Reviewing the development of deep learning, the pursuit of this goal can be divided into two intertwined stages: the first one aims to build and train deeper networks to digest more data; the second one tries to further mine the potential of given data by improving the network’s parameter efficiency. As the depth of the network in computer vision has becomes saturated, research focusing on exploring network redundancy and designing a more compact architecture has received more attention recently. To the best of our knowledge, in biomedical image computing, there is still little systematic effort dedicated to the reduction of the network’s redundancy. The present work relies heavily on the following two aspects of efforts to reduce parameter redundancy in the field of computer vision.
Many works resort to exploring the redundancy of the network in a post-processing manner. Among these efforts, the Low Rank Approximation (LRA) methods are most relevant to ours. By viewing the convolutional layers as high order tensors, these methods compress the convolutional layers of pre-trained networks by finding their appropriate LRA. Using low rank decomposition to accelerate convolution was first suggested by [
13] in codebook learning. In the context of CNNs, the work in [
14] proposed a Canonical Polyadic (CP) decomposition and clustering scheme for the convolutional kernels. Pre-trained 3D filters are approximated by consecutive 1D filters, and the error is minimized by using clustering and post-training. The work in [
15] suggested using different tensor decomposition schemes, and an iterative scheme was employed to get an approximate local solution. The work in [
16] further extended the use of CP decomposition and proposed a different low rank architecture that enabled both approximating an already trained network and training from scratch.
Rather than designing an LRA method, another group of works aimed to improve parameter efficiency. In [
17], a spatial separation of the convolution operator was proposed, where the 3 × 3 kernels were separated into two consecutive kernels of shapes 3 × 1 and 1 × 3. Jin et al. [
18] exploited structural constraints to conventional 3D CNNs (including channel and spatial dimensions) to reduce the computational cost via separable convolutions. To speed up the computation and reduce the model size, Gonda et al. [
19] proposed a novel strategy via replacing 3D convolution layers with pairs of 2D and 1D convolution layers. Numerous other attempts on depthwise separable convolutions have been made in various fashions to improve the efficiency of convolution [
20,
21,
22]. Zhang et al. [
23] combined depthwise separable convolution and spatial separable convolution for liver tumour segmentation. In this paper, we separate 3D kernels into 1D kernels, and
Table 1 shows the comparison with other relevant methods.
3. Methods
We start this section by discussing the separability of 3D convolutional kernels and the issues that may arise. Then, based on the infrastructure provided by the spatial decomposition of kernels, we construct the proposed 3D-DSC module that possesses better parameter efficiency and stronger nonlinear representational power.
3.1. 3D Separability of Convolutional Kernels
Given a volumetric image, when we employ a 3D convolution kernel to generate a 3D feature volume, the input to the network is the entire volumetric data. By leveraging the kernel sharing across all three dimensions, the network can take full advantage of the volumetric contextual information. Generally, the following equation formulates the exploited 3D convolution operation with stride one in an element-wise fashion:
where
is the 3D kernel of size
in the
lth layer, which is connected to the
kth input feature volume
in the previous layer, and the
jth output feature volume
,
is the element-wise value of the 3D convolution kernel. Assume the
lth layer has
K input feature volumes, and let
denote the element-wise nonlinear activation function and
the corresponding bias term; the output feature volume
is obtained as:
Mathematically, the 3D kernel tensor
can be factorized into a linear combination of rank 1 tensors according to the CP decomposition:
where
R is the rank of
, ⊗ denotes the outer product operation and
,
,
are 1D vectors. Element-wise, the above equation can be rewritten as:
Substituting (4) into (1) gives the following equivalent expression for the evaluation of the 3D convolution:
With this formulation, the 3D convolution can be recast as a sequence of 1D convolutions. From inside out, the calculation within the parentheses can be viewed as: first convolve the feature volume with a 1D filter along the X dimension, then followed by the 1D convolution with and along the Y and Z dimension successively. Vectors , and can be viewed as the corresponding horizontal (H), vertical (V) and lateral (L) 1D filters, respectively.
Assuming that the rank of kernel tensor
is equal to one (i.e.,
R = 1), the 3D convolution can be decomposed into a sequence of three 1D convolutions as shown in
Figure 2 (
). Note that convolution is a linear operator, and the 1D filters as shown in
Figure 1 can be arranged in any order.
Rank 1 is a strong assumption, and the intrinsic rank of
is generally higher than one in practice, however, the generalization from the rank 1 topology to the rank R case is straightforward. Equation (
3) shows that the rank R tensor is the sum of R rank 1 tensors, and this suggested that the rank R topology can be constructed by simply concatenating R copies of the rank 1 topology, as shown in
Figure 2 [
24].
3.2. 3D Dense Separated Convolution Module
Although the 3D separated convolution topology described in the previous section is mathematically equivalent to direct 3D convolution, the profits of this decomposition are reflected in the following aspects:
First, the rank constraints of 3D convolution kernels can be easily encoded in the network’s topology by stacking
k (
) groups of horizontal, vertical and lateral (HVL) 1D convolutions (as seen in
Figure 2). Once the model structure is defined, we can leverage the traditional CNN training method to learn more compact weights from scratch, thus avoiding the traditional post-processing stage of applying the low rank constraint on the pre-trained network, then followed by the iterative fine tuning of layers. In addition, the possible information loss and performance degradation caused by low rank constraints can be minimized as a whole upon training. We will show that the precision can even be increased in the Experiments Section.
Second, when a rank k topology is applied to replace the original full rank 3D convolution kernel, the number of independent parameters per-filter can be reduced from to , which results in a significant reduction of the overall learnable parameters for small k considering the huge number of filters deployed in the network. Since the training data size in many biomedical tasks is much smaller than that of computer vision, this reduction in the amount of parameters will reduce the risk of overfitting during training and enable deeper network design.
Finally, the cascaded 1D convolution structure provides the possibility to further improve the nonlinear representation capability of the network. Since the linear combination of convolution operations is still linear, the current decomposed topology can only increase the network’s visual depth, but not the effective depth. However, with this structure, the effective depth of the network can be easily increased by inserting the nonlinear activation layers (e.g., leaky ReLU layers) between the concatenated 1D convolutions, thus increasing the nonlinearity of the network and encouraging the learning of more discriminative features.
However, there are two issues inherited in this kernel decomposition. First, the serialized model with 1D convolutions is more vulnerable to the vanishing gradient problem than standard 3D CNNs. Accompanied by the increase of the network’s depth, longer gradient propagation paths may result in fast gradient decaying, as well as difficulty in optimization. Second, once the nonlinear activation layer is inserted between the 1D filters, the different ordering of the 1D filters will no longer be equivalent.
Inspired by the recent success of densely connected networks [
25], we propose to extend the 3D separated convolution discussed in the previous section by further introducing dense connections between 1D filters.
Figure 1 illustrates the layout of the rank R 3D-DSC module schematically.
Similar to the DenseNet, we introduce direct connections from any layer to all subsequent layers within each block. In order to maximize the information flow, the features are concatenated and then followed by a composited operations including Batch Normalization (BN) and leaky Rectified Linear Units (leaky ReLU) before they are passed to the next layer. Although each 1D decomposed convolution layer has less parameters, it typically has more input feature maps due to the dense concatenation. It was demonstrated that a 1 × 1 convolution can be employed as a bottleneck layer before each 3 × 3 convolution to reduce the number of input channels [
11,
25]. To reduce the parameters of 3D-DSC, in this study, we added a 1 × 1 × 1 convolution in each 1D decomposed convolution. In our implementation, we restricted each layer to produce half the number of feature maps as the input. Assume there are k feature maps in the input layer; the concatenate operation after the last 1D decomposed convolution layer will accumulate the feature map to the number of
. In order to make the number of output feature maps consistent with that of direct 3D convolution, we introduced an additional bottleneck layer consisting 1 × 1 × 1 convolution after the last 1D convolution layer. With this design, the extension from rank 1 3D-DSC to the rank k case will be the same as the naive 3D separated convolution version, as discussed in the previous section, i.e., by simply stacking
k copies of the rank 1 topology.
By introducing the within block dense connections, each 1D kernel is provided with the opportunity to access the input feature map directly, thus to some extent alleviating the ordering problem of 1D kernels. In addition, the employment of dense connections also brings the three following benefits that relieve our previous concerns in a precise manner. First, direct connections between all layers help improve the flow of information and gradients through the network, alleviating the problem of the vanishing gradient. Second, short paths to all the feature maps in the architecture introduce an implicit deep supervision. Third, dense connections have a regularizing effect, and considering the reduction in the number of learnable parameters introduced by 3D separated convolution, such a joint effort would substantially reduce the risk of overfitting under limited training data, which is an essential problem for most biomedical image analysis applications.
Since the size and the number of feature map of our 3D-DSC block are consistent with that of direct 3D convolution, we can directly substitute the 3D convolution layers with 3D-DSC in the existing 2D CNNs and enjoy the benefits of 3D-DSC. If using a high level library such as Keras or TensorFlow-Slim, it would take only several lines of code.
3.3. 3D CNN Architecture Based on 3D-DSC
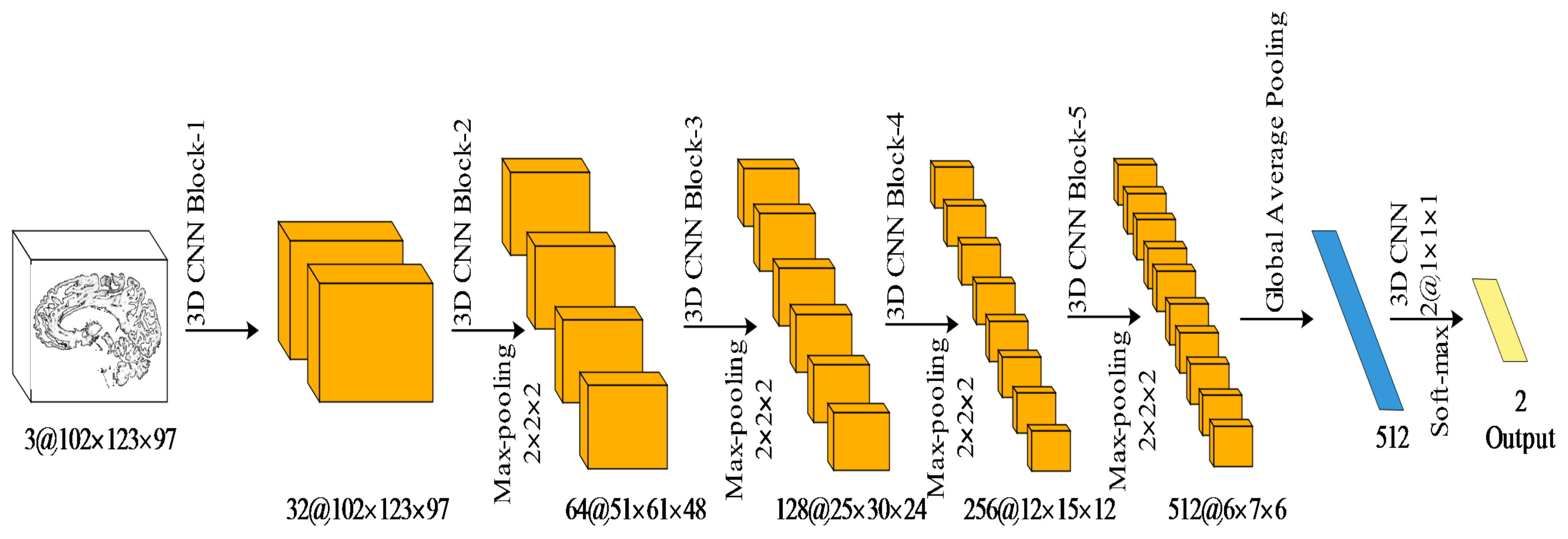
For the classification task, we constructed a simple 3D CNN architecture to diagnose attention deficit hyperactivity disorder.
Figure 3 demonstrates the proposed CNN architecture. The architecture followed the typical design philosophy of a convolutional network. It consisted of the repeated application of the 3D convolutional blocks (
Table 2 shows the number and the type of 3D convolutions in each block), each followed by a
3D max pooling operation with stride 2 for downsampling. After each downsampling layer, we doubled the number of volumetric feature channels. At the final layer, a 3D global average pooling and a
3D convolution were used to map each volumetric feature vector to the desired number of classes.
For the segmentation task, we applied the classic 3D U-net architecture. We kept the typical encoder-decoder structure and the number of blocks in each path. Different from the original 3D U-net, we used instance normalization and leaky ReLUs, rather than batch normalization and ReLUs. Based on these, we designed a universal 3D U-net block with 3D-DSC.
Figure 4 shows the proposed 3D U-net block. The block reserves the first normal 3D convolution, followed by 2 3D-DSC.
3.4. Training of the 3D CNN Architecture
Both classification and segmentation CNNs were trained end-to-end on the datasets of brain scans in MRI. An example of the typical content of such volumetric medical image is shown in
Figure 5.
In this paper, we select the cross-entropy as the classification loss function and the Dice loss as the segmentation loss function. The cross-entropy
can be written as:
where
N is the number of samples,
and
are the input and corresponding label of the
nth sample,
is the function learned by the network and
represents the output of the neural network given the input
. The Dice loss
D for binary classes is defined as follows:
where the sums run over the
N voxels, of the predicted segmentation volume
and the ground truth volume
.
We employed a similar training strategy during classification and segmentation. It is worth noting that adaptive optimization methods have better performance in the early stage of training, but are outperformed by Stochastic Gradient Descent (SGD) at later stages. To minimize the effect of random initialization, we firstly trained the model with random initialization and the Adam [
28] optimizer. Then, we refined the model with the SGD optimizer. The learning rate was initially set to 0.00001 and decreased by a factor of 10 when the validation error stopped decreasing. The early-stopping strategy was used with patience of 50. We denote the mean difference between the training loss and the validation loss within the last 50 epochs as the Overfitting Distance (OD), which can be used to evaluate the ability of the network to cope with overfitting. The OD can be written as:
where
N is the training epoch numbers,
is training loss and
is validation loss. In our experiments, we employed 5 fold cross-validation to evaluate the proposed method.