Abstract
The chemical composition of groundwater and related surface water ecosystems can be modified by intensive agricultural activities. This is the case of the Natural Reserve of Pétrola saline lake (Albacete, SE Spain), which constitutes the discharge area of an unconfined aquifer. The extended use of fertilizers and pesticides poses a threat to ecosystem sustainability. One of the most applied herbicides worldwide has been atrazine. Despite being prohibited in Spain since 2007 by European regulations, atrazine can still be detected due to its high persistence in the environment. Atrazine degradation pathways are mediated by biological processes performed by microorganisms with adapted metabolic mechanisms that make in situ bioremediation possible. To evaluate the presence of such microorganisms in the unconfined aquifer, groundwater was collected from a flowing 37.9 m deep piezometer. DNA was extracted, and the bacterial 16S rRNA gene was amplified and cloned. Later, 93 clones were sequenced, providing the first molecular assessment of bacterial community structure in the deep zones of the aquifer. Some of these bacteria have been previously described to be involved in atrazine degradation. In addition, 14 bacteria were isolated from the groundwater samples and identified by 16S rRNA gene sequencing. DNA from these bacteria was subjected to PCR assays with primers designed for the genes involved in the atrazine degradation pathway. Positive results in the amplification were found in at least three of these bacteria (Arthrobacter sp., Nocardioides sp. and Pseudomonas sp.). The atrazine-degrading genetic potential was shown to be dependent on the trzN and atzA,B,C gene combination. These results suggest for the first time the adaptation of the bacterial population present in deep aquifer zones to atrazine exposure, even after more than 15 years of its ban in Spain. In addition, this study provides the baseline data about the bacterial communities found in deep aquifer zones from the hypersaline lake-aquifer system.
1. Introduction
Contamination of groundwater due to environmentally unsustainable human activities has become one the most important pollution threats worldwide. The main sources of pollutants are derived from industrial and agricultural activities. Thus, many inorganic and organic compounds, including pesticides, have increased their presence in surface and groundwater bodies. Pollutants have the potential to modify groundwater chemical composition as well as groundwater ecosystems [1,2].
Nitrogen-based compounds, such as fertilizers or pesticides, have widespread use in agricultural regions. They may promote eutrophication of surface water bodies and, due to their toxicity, represent a risk for both human health and the survival of aquatic species [3]. Atrazine is within this group of compounds. It is one of the most widely applied herbicides all over the world since its introduction in the 1950s. The persistence of atrazine, its moderate water solubility, and high movement through soil and water key to its potential role as a pollutant in water bodies [4,5,6]. Atrazine concentrations above the allowed limits of 0.1 µg/L, established by the European Union [7], have been found in drinking water [8]. In addition, its disruptor endocrine activity is demonstrated with pernicious potential effects on human [9,10] and animal health [11,12]. As a consequence, atrazine application was prohibited by the European Union in 2004, although in Spain it was extended until 2007. Even after more than 15 years of being banned in EU and Spain, nowadays considerable concentrations of atrazine and its metabolites are still detected in surface and groundwater bodies due to their elevated persistence in the environment [13]. Over the past few decades, researchers’ concerns about atrazine removal paths have been raised regarding its environmentally persistence and water quality impact [14,15,16,17,18].
Biological processes are the predominant atrazine degradation pathways. Shapir et al. [19] revised and explained the enzymes and genes involved in microbial s-triazine metabolism. The reactions occur in an obligate order with funneling of broad-specificity enzymes, encoded by atzA,B,C/trzN genes, into cyanuric acid as an intermediate product. Then, subsequently narrow-specificity enzymes, codified by atzD,E,F/trzD,F genes, finish the s-triazine ring cleavage. The AtzA/TrzN enzymes start a hydrolytic dechlorinating reaction to produce the hydroxyatrazine molecule, followed by a sequential and stepwise alkylamino chains removal carried out by AtzB and AtzC enzymes to produce N-isopropyammelide and cyanuric acid, respectively. Then, AtzD/TrzD enzymes hydrolyze cyanuric acid to a biuret intermediate, which is hydrolyzed to allophanic acid by the AtzE enzyme. Finally, allophanic acid is the single hydrolyzing substrate for the AtzF enzyme to produce carbon oxide and ammonia.
Microbial communities within aquifers are expected to consist largely of heterotrophs and chemoautotrophs well adapted to the nutrient-poor and oligotrophic groundwater environment [20]. Moreover, under an environment with a long history of repeated xenobiotic exposition, microorganisms are expected to adapt their metabolic mechanisms progressively to carry degradative genes for foreign pollutants [21,22,23]. In many deep aquifers, which contain huge water volumes with slow turnover times, in situ bioremediation performed by the local communities of microorganisms is very interesting and sometimes, the only decontamination recourse [24].
This study was carried out to evaluate the presence of bacteria related to atrazine degradation in deep zones of the Pétrola hypersaline lake-aquifer system (southeastern Spain). The first molecular assessment of the groundwater bacterial community structure has been made by the sequencing of 16S rRNA gene clones obtained from groundwater samples collected at 37.9 m deep. In addition, the atrazine-degrading genetic potential is determined in isolated bacteria by PCR in order to assess their possible use in pesticide natural bioremediation processes.
2. Materials and Methods
2.1. Study Area
The study area is one of the main representative saline wetlands in Castilla-La Mancha (SE Spain) and constitutes an important hotspot of biodiversity. As a result, it has many national and international protection statuses: “Sites of Community Importance” EU 1997, “Natural Reserve” Castilla-La Mancha Regional Government (DOCM 26/2003), and Special Protection Area (ES0000153). The hypersaline lake-aquifer system consists of an endorheic basin that extends over 43 km2, where Pétrola Lake, which is connected to the aquifer, occupies the terminal discharge zone. Concerning land uses, farming covers about 75% of the basin area, comprising principally dryland (55%) and irrigation croplands (15%). Agricultural pollutants reach the saline lake through several streams and springs, which constitutes groundwater discharges zones (Figure 1). Regarding atrazine concentrations, previous studies showed values ranging from 0.104 to 0.264 µg/L [25]. Moreover, untreated urban wastewaters are discharged directly into the lake. Despite the fact that is a protected environmentally significant area, the impact of human activities is noticeable. Consequently, the study area was declared as a highly vulnerable zone to nitrate pollution in 1998 and deemed as a heavily modified water body by the competent water authorities.
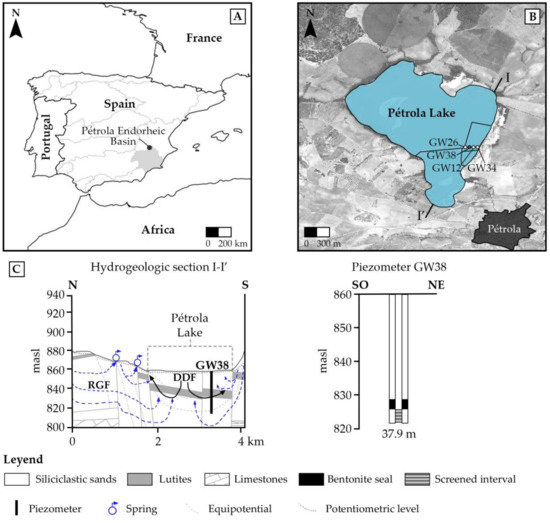
Figure 1.
(A) Location of the Pétrola endorheic basin in southeast Spain. (B) Location of piezometers installed in Pétrola Lake. The piezometer which withdraws regional groundwater was selected for groundwater sampling. (C) Simplified hydrogeological section showing the main groundwater flow components and the allocation of the piezometer GW38. In the same caption is shown the building characteristics of the piezometer GW38 with an indication of the screening zone and sealed interval. masl: meters above sea level. DDF: density-driven flow. RGF: regional groundwater flow.
A complete hydrogeological description can be found in Gómez-Alday et al. [26]. Briefly, the system has two main flow components: a regional groundwater flow, from the recharge to the discharge lake area, and a density-driven flow, conducted by differences in densities between the saline surface water (up to 1.23 dS/cm; November 2010) and groundwater [27]. During 2008, a set of four PVC piezometers were installed in a dike near the eastern bank of the lake to study surface water and groundwater interactions. The PVC piezometers were 5 cm in inner diameter and were installed at different depths: 12.1 m (GW12), 25.8 m (GW26), 34.1 m (GW34), and 37.9 m (GW38). Screening zones were isolated by means of internal bentonite seals. In this study, the deepest piezometer (GW38, 38°50′17.6337″ N, –1°33′33.9167″ W) was selected for studying the microbial community and atrazine-degrading potential in the deep aquifer zones (Figure 1). The flowing piezometer allows for the collecting of samples from the regional groundwater flow. Published tritium values suggest groundwater recharge occurred before the 50s decade, corresponding to long-term resident groundwater [26]. This piezometer was selected to be representative of the deep aquifer environmental conditions for studying its indigenous bacterial community structure.
2.2. Groundwater Sampling
The groundwater sample (3 L) was collected from a flowing piezometer (GW38) screened at a depth of 37.9 m in March 2019. Physico-chemical parameters were measured in situ using an HQ40d portable Multi-Parameter Meter (Hach Company, USA). The accuracy of physico-chemical determinations was ±0.3 °C for temperature, ±0.002 for pH, ±0.5% for electrical conductivity (EC), ±200 mS/cm for total dissolved solids (TDS), ±0.1 mV for Eh, and ±0.01 mg/L for dissolved oxygen. Atrazine concentrations were analyzed by gas chromatography coupled to a mass detector in an Agilent 7890B Series Gas Chromatograph linked to an Agilent 5977B Mass Selective Detector equipped with a DB-5ms capillary column (J&W Scientific, USA). Physico-chemical parameters and atrazine concentration of the groundwater from this piezometer have belonged to a paired monitoring study since April 2018, with monthly measurements. Before groundwater sampling, the well was drained for 45 min to collect a representative sample from the regional groundwater. Afterward, 3 L of groundwater were collected in a sterile and pre-rinsed polyethylene bottle. The recipient was sealed with a screw cap, avoiding headspace, refrigerated in darkness, and transported to the laboratory not exceeding five hours. At the laboratory, groundwater samples were immediately processed.
2.3. DNA Extraction
Environmental DNA extraction was performed using the NucleoSpin® Soil DNA extraction kit (Macherey-Nagel, Düren, Germany) according to the manufacturer’s instructions with slight adaptation for water samples and bacteria. After homogenization, the groundwater sample (3 L) was filtered throughout nylon membranes with a 0.45 µm pore diameter (Merck KGaA, Darmstadt, Germany), at a rate of 1 nylon membrane per 500 mL, approximately. The membranes were cut into small fragments, immersed into lysis buffer, and vortexed. The resultant supernatant was taken to further proceed to DNA extraction following the manufacturer’s instructions.
Regarding bacteria DNA extraction, 3 mL of each bacterial culture was centrifuged. The resultant pellet was resuspended in lysis buffer and used to further proceed to DNA extraction. The yield and quality of extracted DNA were quantified by 1% agarose gel and spectrophotometer, using a multimode lector Cytation 5 with the specific software Gen5 (Biotek, Vermont, USA) at the Biotechnology and Natural Resources Laboratory (Institute for Regional Development, University of Castilla-La Mancha, Spain).
2.4. Cloning
The 16S rRNA gene fragment was amplified from 100 ng of groundwater sample DNA in a 25 µL reaction volume containing 1U of DNA polymerase (Promega, Madrid, Spain), 1x reaction buffer, 4 mM MgCl2, 0.4 mM dNTP, and 0.25 mM of 16S rRNA gene primers listed in Table 1. DNA amplification was carried out in a Gene Pro thermocycler (Bioer Technology Co., Zhejiang, China) under the following conditions: 4 min at 94 °C, 35 cycles of 1 min at 94 °C, 1 min at 53 °C, 1 min at 72 °C, plus an additional 4 min-cycle at 72 °C. Next, 16S rRNA gene amplicons were checked in agarose gel and purified to clone them into the pGEM-T® Easy Vector System (Promega, Fitchburg, WI, USA) according to the manufacturer’s instructions.

Table 1.
Sequence and conditions of primer pairs used in this study.
2.5. Strain Isolation
Direct spreads of 100 µL of groundwater samples were carried out in six sterile LB plates and were incubated at room temperature for five days in a laminar flow hood. Visually different colonies were isolated in 1 mL of LB culture to grow for 24 h. Then, 100 µL of each culture of isolated bacteria was spread individually in plates, replicating them successively to obtain a pure isolated culture. Isolated strains were preserved in 50% glycerol and kept frozen at −80 °C for further analyses.
2.6. Sequencing of the 16S rRNA Gene Fragment
Sequencing of the 16S rRNA gene fragment from recombined plasmids and 16S rRNA gene fragments from each isolated bacterium was done in an Applied Biosystems ABI 3730xl DNA Analyzer by Macrogen (Madrid, Spain). These sequences were trimmed to avoid sequencing errors, plasmid sequences, and low-quality ends. The 16S rRNA gene sequences from both cloning and isolated bacteria were deposited in the GenBank database under the accession numbers MT941683-MT941753 and MT815708-MT815721, respectively. Sequences were compared with the 16S rRNA gene library bacteria and archaea of the GenBank using the BLASTN tool [28].
2.7. PCR Assays of Atrazine-Degrading Genes
The composition of the atrazine-degrading genetic potential of each isolated strain was determined by PCR using primers and optimized annealing temperatures listed in Table 1. Approximately, 50 ng of bacterial DNA was used as a template in PCR assays. PCR reactions were carried out in 25 µL reaction volume containing 1U of DNA polymerase (Promega, Madrid, Spain), 1x reaction buffer, 4 mM MgCl2, 0.4 mM dNTP, and 0.25 mM of each primer pair. Amplification conditions were 10 min at 94 °C, 35 cycles of 0.5 min at 94 °C, 0.5 min at considered annealing temperature (Table 1), 0.5 min at 72 °C, plus an additional 10 min-cycle at 72 °C and 7 min-cycle at 4 °C. All PCR reactions were performed in duplicate.
3. Results
3.1. Physico-Chemical Parameters of Deep Groundwater
Groundwater from the 37.9 m deep piezometer has been monitored since April 2018, and physico-chemical data have been collected monthly. These data (as well as data collected from other points) have been used to characterize the hydrogeological characteristics of the lake, which have been published in previous studies [34,35,36]. Basically, between April 2018 and November 2019 the average values for physico-chemical parameters of groundwater collected from the 39.8 m deep piezometer were 7.45 ± 0.16 for pH, 16.57 ± 2.17 °C for temperature, 2.17 ± 1.00 mg/L for dissolved oxygen, and 23.22 ± 0.09% for oxygen saturation, 28.61 ± 38.85 mV for Eh, 2315.50 ± 172.02 µS/cm for EC, and 1436.44 ± 140.23 mg/L for TDS. Concerning atrazine concentrations varied from below the detection limit up to 224.43 ng/L (July 2019). Thus, deep groundwater is circumneutral, brackish, and near to reducing redox conditions.
3.2. Structure of the Groundwater Microbial Community
Sequencing of clones, which contained 16S rRNA gene inserts from the groundwater sample, resulted in a total of 93 sequences. After checking their quality and end-trimming, the sequences were identified by comparisons with bacteria and archaea libraries from NCBI, according to the threshold of 95% and 98.65% of similarity for genus and species identification, respectively [37,38]. No archaeal clones were obtained. This study constitutes a first view of the most abundant bacteria that inhabit the groundwater environment of the Pétrola lake-aquifer system. It is far from being considered a complete characterization of the bacterial community, which must be done with other modern techniques such as next-generation sequencing. However, the results obtained show the presence of certain bacteria that have been reported to be involved in natural bioremediation processes.
The sequences clustered into two phyla, Proteobacteria and Firmicutes, with 97.8% and 2.2% of relative abundance, respectively. Within Proteobacteria, the subphylum Rhodobacteria is completely represented with its members of Gammaproteobacteria, Betaproteobacteria, and Alphaproteobacteria classes, covering 49.4%, 29.2%, and 19.1% of relative abundance, respectively. In contrast, within Firmicutes only the class Bacilli is identified with 2.2% representation.
The comparison of the sequences from the 93 clones resulted in 36 species identified, which were clustered into 18 genera and 10 families, listed in Table 2. At the family level, the dominant taxa were Pseudomonadaceae and Comamonadaceae, covering together a 58.4% of the global abundance. These families, which together with Caulobacteraceae, Xanthomonadaceae, Bacillaceae, and Rhizobiaceae accounted for 69.7% of relative abundance over the total sequences analyzed, are closely reported to nitrogen cycling, including the nitrate removal by denitrification processes [39,40,41,42,43]. Chromatiaceae and Rhodobacteraceae were found to cover 11.2% and may constitute a sulfur turnover consortium. The members of these families were found to be deeply involved in sulfur and carbon biogeochemical cycling, with competitive advantages in sulfur metabolic mechanisms, and symbiosis with aquatic micro- and macroorganisms [44,45]. In addition to carbon cycling, Sphingomonadaceae, Moraxellaceae, and some specific genera (i.e., Phenylobacterium) have enzyme specificity and kinetics involved in the degradation of a wide range of xenobiotics, which can include pesticides such as atrazine [46,47,48]. At the genus level, Pseudomonas was the most abundant Proteobacteria found in the groundwater sample analyzed, accounting for 29.2% of the total bacterial sequences in the sample with 11 different related species. Also, genera from the above-mentioned consortium of families with xenobiotic abilities, Sphingopyxis, Acinetobacter, and Phenylobacterium, showed eight different species, becoming the third most abundant group.

Table 2.
Putative classification and relative abundance of bacteria from groundwater.
3.3. Isolation of Groundwater Bacteria Strain
Regarding the bacteria isolation carried out in this study, up to 45 visually different colonies were individually grown in tubes and spread-replicated into LB plates until a pure culture was obtained. Then, 16S rRNA sequencing revealed 14 different strains clustered into 11 genera, 8 families, and 3 phyla (with one single class each phylum) listed in Table 3. Classes Actinobacteria (Actinobacteria), Bacilli (Firmicutes), and Gammaproteobacteria (Proteobacteria) showed a contribution of 62.5%, 25%, and 12.5%, respectively. The dominant family taxon was Microbacteriaceae with 42.9% of relative abundance, followed by Micrococcaceae with 23.9%. Primarily, Microbacteriaceae, along with Dietziaceae, Nocardioidaceae, and Intrasporangiaceae, which constituted a consortium of 62% relative abundance, have been reported to contain enzymatic mechanisms related to the degradation of recalcitrant xenobiotics compounds from the environment [46,49,50,51,52], as well as hydrocarbons, sugars, and other micro- and macroorganisms by saprophytic methods [53,54]. On the other hand, Micrococcaceae along with Bacillaceae and Pseudomonadaceae, form a clade closely linked to nitrogen cycling. Some members of these families can perform denitrification processes commonly by nitrate-reduction or by degrading nitroaromatic pesticides, such as atrazine [50,55]. The adaptation ability of some members of the genera, Arthrobacter, Pseudomonas, Nocardioides, and Sphingomonadaceae, and their well-adapted mechanisms to process atrazine are well known and largely demonstrated [8].

Table 3.
Classification and alignment hit table of isolated strains from groundwater.
3.4. Atrazine Degrading Genetic Potential of Groundwater Bacterial Isolates
Atrazine-degrading genetic potential of each isolated bacteria was determined by PCR screening for the specific genes atzA,B,C,D,E,F and trzN,D coding for enzymes responsible for atrazine transformation (Figure 2). Amplification by PCR allows for the estimation of the presence/absence of the genes involved in the atrazine degradation metabolic pathway, which gives an idea of the genetic potential of groundwater or isolated bacteria to carry out this pathway. The results of the PCR amplification assays using groundwater community DNA as a template revealed the presence of atzA, atzB, atzC, atzE, atzF, trzN, and trzD sequences (see Table 4). No PCR products were obtained for the atzD gene after electrophoresis. Eight of the fourteen isolated strains may possess atrazine-degrading genetic potential activity.
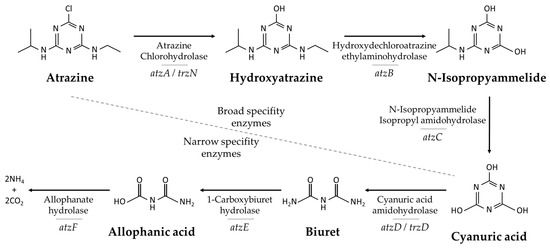
Figure 2.
Atrazine degradation pathway. Each reaction shows the genes screened and the name of the corresponding enzyme responsible for catalyzing the reaction.

Table 4.
Atrazine degrading genes present in isolated bacteria. +: positive amplification signal; -: no amplification signal.
PCR amplification of trzN and atzA genes showed positive results in a higher number of isolates than the rest of the degrading gene-coding enzymes, while atzB and atzE were amplified only in a single microorganism. Both genes –trzN and atzA– and competent strains, initiated the degrading pathway by a hydrolytic dechlorinating reaction leading to a non-chlorinated more biodegradable s-triazine. TrzN was the most detected gene, since five strains –GW38-4, GW38-6, GW38-7, GW38-10, and GW38-14– associated with Microbacterium, Arthrobacter, Nesterenkonia, Nocardioides, and Pseudomonas, showed its presence. Strains GW38-3, GW38-6, and GW38-10 close to Microbacterium, Arthrobacter, and Nocardioides, also showed potential genetic activity in atzA. AtzB and atzE genes were only amplified by the Pseudomonas related strain, GW38-14. Some strains of Microbacterium appeared to be responsible for atzC signal. Instead of atzD gene existence, signals were found for its enzyme homolog trzD by strains GW38-5 and GW38-7 related to Labedella and Nesterenkonia. Finally, only strains GW38-7 and GW38-10, closest to Nesterenkonia and Nocardioides, showed positive amplification for atzF coding enzyme probes.
4. Discussion
As a first approach to evaluate the presence of bacteria related to atrazine degradation processes, DNA was extracted from groundwater and the 16SrRNA gene was amplified and cloned. Subsequently, 93 clones were partially sequenced and compared with 16SrRNA databases to identify the bacteria. This is a quick and easy method to gain a view of the most abundant bacteria present in groundwater, although the information obtained is far from providing a complete characterization of the structure of the bacterial communities that inhabit groundwater, due to the low number of clones analyzed. In this sense, a higher number of clones sequenced from the sample would have resulted in greater species diversity. However, despite the low number of clones sequenced, it was possible to find bacteria that have been described to be involved in these bioremediation processes. Thus, the sequencing of 93 clones from deep groundwater in the Pétrola lake-aquifer system must be enough to find the predominant bacteria and provided information about the possible biogeochemical processes carried out by microorganisms adapted to low oxygen and brackish groundwater ecosystems. This constituted the first approach to the knowledge of bacterial communities adapted to the deep zones of a hypersaline lake-aquifer system, in which environmental conditions may differ substantially from aquatic surface environments.
The hypersaline lake-aquifer system may constitute a natural barrier to organic pollutants promoted by the existence of adequate biogeochemical conditions. It is noticeable the presence in deep groundwater zones of the aquifer of microorganisms belonging to families such as Pseudomonadaceae, Comamonadaceae, Xanthomonadaceae, Bacillaceae, or Rhizobiaceae (69.7%), of which some of their members have been found related to nitrogen compounds degradation, including the banned herbicide atrazine in EU. Additionally, it is remarkable the presence of bacteria from families such as Sphingomonadaceae, Moraxellaceae, or the Phenylobacterium genus from the Caulobacteraceae family (21.4%), which have been associated with mono- and multifunctional microorganisms related to xenobiotic compounds degradation. The transport and accumulation of contaminants in the groundwater derived from anthropic activities can cause processes of metabolism adaptation in the microbial community [56].
Regarding atrazine biodegradation, it has been reported to be a slow process due to its asymmetric substituent groups [57]. Nowadays, it is known that the acceleration of the biodegradation process is mainly due to the enzymatic-degrading pathway dispersed among a wide range of microorganisms. As a response to prolonged exposure of atrazine, microbial communities might be able to adapt to metabolize the compound, facilitating the biodegradation and thereby reducing toxicity in the contaminated sites [58,59]. The microbial degradation pathways play the primary role in the reduction of toxic chemicals from contaminated aquatic environments. It has been described that atrazine catabolic pathways appear to be the result of the recruitment of different genes in broad microbial communities rather than a genetic element encoding the entire pathway. The predominance of the Pseudomonas strain clones found in the analyzed groundwater sample suggests that their members could play a role in atrazine degradation within the aquifer bacterial community since it has been reported that some Pseudomonas sp. strains are able to use atrazine as a nutrient source [50]. On the other hand, the members of the Sphingomonadaceae family have been described for their ability to adapt more rapidly by harboring different pathways for metabolic degradation of new compounds in the environment than members of other bacterial genera [48].
This report provides the first approach of the atrazine-degrading genetic potential of fourteen cultivable bacteria strains isolated from the regional groundwater flow. The characterization of atrazine-degrading bacterial strains revealed eight strains linked to the existence of atzA,B,C,E,F and trzN,D genes, which encode the enzymes responsible for the mineralization of this herbicide. Five of the eight newly isolated strains possessed the trzN gene. This high proportion of bacteria possessing the trzN gene, which encodes an atrazine chlorohydrolase, shows its widespread occurrence throughout the potential atrazine-degrading bacterial community [60] and its prevalence over the atzA gene. Seven strains out of eight harbored the trzN/atzA–atzB,C gene combination. This atrazine-degrading gene combination leads to the transformation of atrazine to its intermediate cyanuric acid. The results, therefore, suggest that the trzN/atzA–atzB,C gene composition may be widespread and highly conserved in different microbial genera. Moreover, such gene combination redundancy suggests an established atrazine-degrading consortium with a cooperative catabolic pathway [61]. By contrast, three of the eight isolated strains showed positive results for the trzD–atzE,F gene combination. The detection of trzN, atzC, and trzD might be an indicator of the fact that atrazine is being degraded via the hydroxyatrazine pathway (trzN) to cyanuric acid (atzC), then to biuret, NH3, and CO2 (trzD). However, the absence of certain genes in the isolated strains, such as atzD, does not mean that they are not present in the whole groundwater environment. Furthermore, other similar genes involved in the missing steps (or unknown genes with the potential to perform the missing steps) were not detected by the primers used in this study, but they could be present. Therefore, atrazine catabolic pathways appear to be the result of the recruitment of different genes in broad microbial communities rather than a genetic element encoding the entire pathway [32].
The presence of bacteria such as Arthrobacter sp., Pseudomonas sp., or Nocardioides sp., that have been reported to contain atrazine degradation genes [18,31,60,61], suggest the potential bioremediation adaptation of the groundwater microbial community. This possible atrazine-degrading capacity and microbial community adaptation must be assessed in future experiments. This study highlights the importance of the use of molecular tools to find possible candidates with the genetic potential to perform natural bioremediation processes, although the details of the expression of these genes, and the real degradative capacity should be verified with further research.
5. Conclusions
This work provides insights into the overall structure of the main bacterial communities and atrazine-degrading genetic potential present in the deep zones of the Pétrola hypersaline lake-aquifer system. The results obtained are a first approach that shows the presence of bacteria clusters that could be related to agricultural-derived pollutant degradation. Nonetheless, subsequent detailed studies of aquifer microbial communities are necessary to further evaluate their functional biodegradation potential. Initial observations of the microbial community structure and isolated bacteria that inhabit the deep groundwater zones of the aquifer suggest that they may be undergoing a natural process of contaminant attenuation. Based on data regarding the presence of genes related to atrazine biodegradation pathways, the isolated strains from the genera Arthrobacter, Pseudomonas, and Nocardioides are the best candidates to test their potential to degrade atrazine in the ecosystem.
Knowledge about the relationship between microbial community structure and environmental conditions in hydrogeological systems is required to develop tools for the assessment and monitoring of natural attenuation processes. Further studies must be focused on the evaluation of the atrazine degrading capacity of the isolated candidates to evaluate the possibility of their use in induced bioremediation technologies.
Author Contributions
Conceptualization, Y.E., M.Á.-O. and J.J.G.-A.; methodology, Y.E., G.A. and J.J.G.-A.; writing—original draft preparation, M.Á.-O. and Y.E.; writing—review and editing, M.Á.-O., Y.E. and J.J.G.-A.; funding acquisition, J.J.G.-A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by a contract (2019-CON-9523) from the Spanish Government-FSE, the CICYT-CGL2017-87216-C4-2-R project from the Spanish government FEDER—Ministerio de Ciencia e Innovación y Universidades-Agencia Estatal de Investigación, and the SBPLY/17/180501/000296L project from Castilla-La Mancha Regional Government.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Böhlke, J.K. Groundwater recharge and agricultural contamination. Hydrogeol. J. 2002, 10, 153–179. [Google Scholar] [CrossRef]
- Elhatip, H.; Afşin, M.; Kusçu, I.; Dirik, K.; Kurmaç, Y.; Kavurmaçı, M. Influences of human activities and agriculture on groundwater quality of Kayseri-Incesu-Dokuzpınar springs, central Anatolian part of Turkey. Environ. Geol. 2003, 44, 490–494. [Google Scholar] [CrossRef]
- Sánchez, J.; Sanabria, J. Metabolismos microbianos involucrados en procesos avanzados para la remoción de nitrógeno, una revision prospectiva. Rev. Colomb. Biotecnol. 2009, 11, 114–124. [Google Scholar]
- Langenbach, T. Persistence and bioaccumulation of persistent organic pollutants (POPs). In Applied Bioremediation—Active and Passive Approaches; Patil, Y.B., Rao, P., Eds.; IntechOpen: London, UK, 2013; pp. 305–329. ISBN 978-953-51-1200-6. [Google Scholar] [CrossRef]
- Graymore, M.; Stagnitti, F.; Allinson, G. Impacts of atrazine in aquatic ecosystems. Environ. Int. 2001, 26, 483–495. [Google Scholar] [CrossRef]
- Funari, E.; Brambilla, A.L.; Camoni, I.; Canuti, A.; Cavallaro, A.; Chierici, S.; Cialella, G.; Donati, G.; Jaforte, A.; Prandi, L. Extensive atrazine pollution of drinking water in the Lombardia region and related public health aspects. Biomed. Environ. Sci. 1988, 1, 350–355. [Google Scholar]
- Council of the European Union. Council Directive 80/778/EEC of 15 July 1980 relating to the quality of water intended for human consumption. Off. J. Eur. Communities 1980, L229, 11–29. [Google Scholar]
- Getenga, Z.; Dörfler, U.; Iwobi, A.; Schmid, M.; Schroll, R. Atrazine and terbuthylazine mineralization by an Arthrobacter sp. isolated from a sugarcane-cultivated soil in Kenya. Chemosphere 2009, 77, 534–539. [Google Scholar] [CrossRef]
- Sass, J.B.; Colangelo, A. European Union bans atrazine, while the United States negotiates continued use. Int. J. Occup. Environ. Health 2006, 12, 260–267. [Google Scholar] [CrossRef]
- Van Leeuwen, J.A.; Waltner-Toews, D.; Abernathy, T.; Smit, B.; Shoukri, M. Associations between stomach cancer incidence and drinking water contamination with atrazine and nitrate in Ontario (Canada) agroecosystems, 1987–1991. Int. J. Epidemiol. 1999, 28, 836–840. [Google Scholar] [CrossRef]
- Vimal, D.; Saini, S.; Kristipati, R.R.; Chowdhuri, D.K. Atrazine or bisphenol a mediated negative modulation of mismatch repair gene, mlh1 leads to defective oogenesis and reduced female fertility in Drosophila melanogaster. Chemosphere 2019, 225, 247–258. [Google Scholar] [CrossRef]
- Hayes, T.B.; Khoury, V.; Narayan, A.; Nazir, M.; Park, A.; Brown, T.; Adame, L.; Chan, E.; Buchholz, D.; Stueve, T.; et al. Atrazine induces complete feminization and chemical castration in male African clawed frogs (Xenopus laevis). Proc. Natl. Acad. Sci. USA 2010, 107, 4612–4617. [Google Scholar] [CrossRef] [PubMed]
- Menchén, A.; Espín, Y.; Valiente, N.; Toledo, B.; Álvarez-Ortí, M.; Gómez-Alday, J.J. Distribution of Endocrine Disruptor Chemicals and Bacteria in Saline Pétrola Lake (Albacete, SE Spain) Protected Area is Strongly Linked to Land Use. Appl. Sci. 2020, 10, 4017. [Google Scholar] [CrossRef]
- Kamaz, M.; Jones, S.M.; Qian, X.; Watts, M.J.; Zhang, W.; Wickramasinghe, S.R. Atrazine Removal from Municipal Wastewater Using a Membrane Bioreactor. Int. J. Environ. Res. Public Health 2020, 17, 2567. [Google Scholar] [CrossRef] [PubMed]
- Ehrl, B.; Gharasoo, M.; Elsner, M. Isotope Fractionation Pinpoints Membrane Permeability as a Barrier to Atrazine Biodegradation in Gram-negative Polaromonas sp. Nea-C. Environ. Sci. Technol. 2018, 52, 4137–4144. [Google Scholar] [CrossRef] [PubMed]
- Khatoon, H.; Rai, J.P.N. Augmentation of Atrazine biodegradation by two Bacilli immobilized on α-Fe2O3 magnetic nanoparticles. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef]
- Ji, Y.; Dong, C.; Kong, D.; Lu, J.; Zhou, Q. Heat-activated persulfate oxidation of atrazine: Implications for remediation of groundwater contaminated by herbicides. Chem. Eng. J. 2015, 263, 45–54. [Google Scholar] [CrossRef]
- Wang, Q.; Xie, S.; Hu, R. Bioaugmentation with Arthrobacter sp. strain DAT1 for remediation of heavily atrazine-contaminated soil. Int. Biodeterior. Biodegrad. 2013, 77, 63–67. [Google Scholar] [CrossRef]
- Shapir, N.; Mongodin, E.F.; Sadowsky, M.J.; Daugherty, S.C.; Nelson, K.E.; Wackett, L.P. Evolution of Catabolic Pathways: Genomic Insights into Microbial s-Triazine Metabolism. J. Bacteriol. 2006, 189, 674–682. [Google Scholar] [CrossRef] [PubMed]
- Griebler, C.; Lueders, T. Microbial biodiversity in groundwater ecosystems. Freshw. Biol. 2009, 54, 649–677. [Google Scholar] [CrossRef]
- Espín, Y.; Sanz, G.; Valiente, N.; Menchén, A.; Toledo, B.; Álvarez-Ortí, M.; Seseña, S.; Gómez-Alday, J.J. The influence of land use on nitrogen and sulfur turnover: A microbial approach. E3S Web Conf. 2019, 98, 06004. [Google Scholar] [CrossRef]
- Van der Meer, J.R. Environmental pollution promotes selection of microbial degradation pathways. Front. Ecol. Environ. 2006, 4, 35–42. [Google Scholar] [CrossRef]
- Moorman, T.B. Adaptation of Microorganisms in Subsurface Environments. In Enhanced Biodegradation of Pesticides in the Environment; Racke, K.D., Coats, J.R., Eds.; ACS Symposium Series: Washintong, DC, USA, 1990; pp. 167–180. ISBN 978-084-12-1784-3. [Google Scholar] [CrossRef]
- Hazen, T.C.; Jiménez, L.; López de Victoria, G.; Fliermans, C.B. Comparison of bacteria from deep subsurface sediment and adjacent groundwater. Microb. Ecol. 1991, 22, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Menchen, A.; Valiente, N.; Sanz, D.; Gómez-Alday, J.J. Atrazine and desethylatrazine distribution in the natural reserve of Pétrola saline lake after 12 years of prohibition. In Proceedings of the 46 Annual Congress of the Internaonal Associaon of Hydrogeologists (IAH2019), Málaga, Spain, 22–27 September 2019; Gómez-Hernández, J.J., Andreo-Navarro, B., Eds.; Asociación Internacional de Hidrogeólogos—Grupo Español: Barcelona, Spain, 2019. [Google Scholar]
- Gómez-Alday, J.J.; Carrey, R.; Valiente, N.; Otero, N.; Soler, A.; Ayora, C.; Sanz, D.; Muñoz-Martín, A.; Castaño, S.; Recio, C.; et al. Denitrification in a hypersaline lake–aquifer system (Pétrola Basin, Central Spain): The role of recent organic matter and Cretaceous organic rich sediments. Sci. Total Environ. 2014, 497–498, 594–606. [Google Scholar] [CrossRef] [PubMed]
- Dountcheva, I.; Doña, C.; Sanz, D.; Sánchez, J.M.; Gómez-Alday, J.J. Quantitative approach to water balance in groundwater discharge hypersaline wetlands. Pétrola Lake, SE Spain. In Proceedings of the 46 Annual Congress of the Internaonal Associaon of Hydrogeologists (IAH2019), Málaga, Spain, 22–27 September 2019; Gómez-Hernández, J.J., Andreo-Navarro, B., Eds.; Asociación Internacional de Hidrogeólogos—Grupo Español: Barcelona, Spain, 2019. [Google Scholar]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000, 7, 203–214. [Google Scholar] [CrossRef] [PubMed]
- De Souza, M.L.; Seffernick, J.; Martinez, B.; Sadowsky, M.J.; Wackett, L.P. The atrazine catabolism genes atzABC are widespread and highly conserved. J. Bacteriol. 1998, 180, 1951–1954. [Google Scholar] [CrossRef] [PubMed]
- Devers, M.; Henry, S.; Hartmann, A.; Martin-Laurent, F. Horizontal gene transfer of atrazine-degrading genes (atz) from Agrobacterium tumefaciens St96-4 pADP1::Tn5 to bacteria of maize-cultivated soil. Pest Manag. Sci. 2004, 61, 870–880. [Google Scholar] [CrossRef] [PubMed]
- Mulbry, W.W.; Zhu, H.; Nour, S.M.; Topp, E. The triazine hydrolase gene trzN from Nocardioides sp. strain C190: Cloning and construction of gene-specific primers. FEMS Microbiol. Lett. 2002, 206, 75–79. [Google Scholar] [CrossRef]
- Rousseaux, S.; Hartmann, A.; Soulas, G. Isolation and characterisation of new Gram-negative and Gram-positive atrazine degrading bacteria from different French soils. FEMS Microbiol. Ecol. 2001, 36, 211–222. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef]
- Valiente, N.; Sanz, D.; Gómez-Alday, J.J. Groundwater recharge by high-salinity lake water in a density-driven flow dominated system: An isotopic approach. E3S Web Conf. 2019, 98, 12024. [Google Scholar] [CrossRef]
- Valiente, N.; Carrey, R.; Otero, N.; Gutierrez-Villanueva, M.A.; Soler, A.; Sanz, D.; Castaño, S.; Gómez-Alday, J.J. Tracing sulfate recycling in the hypersaline Pétrola Lake (SE Spain): A combined isotopic and microbiological approach. Chem. Geol. 2017, 473, 74–89. [Google Scholar] [CrossRef]
- Espín, Y. Caracterización de la diversidad bacteriana en la laguna hipersalina de Pétrola. Master’s Thesis, University of Castilla-La Mancha, Albacete, Spain, September 2019. [Google Scholar]
- Tindall, B.J.; Rosselló-Móra, R.; Busse, H.J.; Ludwig, W.; Kämpfer, P. Notes on the characterization of prokaryote strains for taxonomic purposes. Int. J. Syst. Evol. Microbiol. 2010, 60, 249–266. [Google Scholar] [CrossRef] [PubMed]
- Stackebrandt, E.; Elbers, J. Taxonomic parameters revisited: Tarnished gold standards. Microbiol. Today 2006, 33, 152–155. [Google Scholar]
- Von Neubeck, M.; Huptas, C.; Glück, C.; Krewinkel, M.; Stoeckel, M.; Stressler, T.; Fischer, L.; Hinrichs, J.; Scherer, S.; Wenning, M. Pseudomonas lactis sp. nov. and Pseudomonas paralactis sp. nov., isolated from bovine raw milk. Int. J. Syst. Evol. Microbiol. 2017, 67, 1656–1664. [Google Scholar] [CrossRef] [PubMed]
- Jayamani, I.; Cupples, A.M. Stable Isotope Probing and High-Throughput Sequencing Implicate Xanthomonadaceae and Rhodocyclaceae in Ethylbenzene Degradation. Environ. Eng. Sci. 2015, 32, 240–249. [Google Scholar] [CrossRef]
- Feng, W.W.; Liu, J.F.; Gu, J.D.; Mu, B.Z. Nitrate-reducing community in production water of three oil reservoirs and their responses to different carbon sources revealed by nitrate-reductase encoding gene (napA). Int. Biodeterior. Biodegrad. 2011, 65, 1081–1086. [Google Scholar] [CrossRef]
- Oh, Y.S.; Roh, D.H. Phenylobacterium muchangponense sp. nov., isolated from beach soil, and emended description of the genus Phenylobacterium. Int. J. Syst. Evol. Microbiol. 2011, 62, 977–983. [Google Scholar] [CrossRef]
- Willems, A.; De Ley, J.; Gillis, M.; Kersters, K. NOTES: Comamonadaceae, a New Family Encompassing the Acidovorans rRNA Complex, Including Variovorax paradoxus gen. nov., comb. nov., for Alcaligenes paradoxus (Davis 1969). Int. J. Syst. Bacteriol. 1991, 41, 445–450. [Google Scholar] [CrossRef]
- Pujalte, M.J.; Lucena, T.; Ruvira, M.A.; Arahal, D.R.; Macián, M.C. The Family Rhodobacteraceae. In The Prokaryotes; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 439–512. [Google Scholar] [CrossRef]
- Pfennig, N.; Trüper, H.G. Isolation of Members of the Families Chromatiaceae and Chlorobiaceae. In The Prokaryotes; Starr, M.P., Stolp, H., Trüper, H.G., Balows, A., Schlegel, H.G., Eds.; Springer: Berlin/Heidelberg, Germany, 1992; pp. 279–289. [Google Scholar] [CrossRef]
- Gaytán, I.; Sánchez-Reyes, A.; Burelo, M.; Vargas-Suárez, M.; Liachko, I.; Press, M.; Sullivan, S.; Cruz-Gómez, M.J.; Loza-Tavera, H. Degradación del poliuretano recalcitrante y aditivos xenobióticos por una comunidad microbiana de vertederos seleccionada y su potencial biodegradativo revelado por análisis metagenómico basado en ligadura de proximidad. Front. Microbiol. 2019, 10, 2986. [Google Scholar] [CrossRef]
- Oh, S.; Choi, D. Microbial Community Enhances Biodegradation of Bisphenol A Through Selection of Sphingomonadaceae. Microb. Ecol. 2018. [Google Scholar] [CrossRef]
- Basta, T.; Keck, A.; Klein, J.; Stolz, A. Detection and Characterization of Conjugative Degradative Plasmids in Xenobiotic-Degrading Sphingomonas Strains. J. Bacteriol. 2004, 186, 3862–3872. [Google Scholar] [CrossRef] [PubMed]
- DeBruyn, J.M.; Mead, T.J.; Sayler, G.S. Horizontal transfer of PAH catabolism genes in Mycobacterium: Evidence from comparative genomics and isolated pyrene-degrading bacteria. Environ. Sci. Technol. 2012, 46, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Singh, A.; Ward, O.P. Biodegradation of nitroaromatics and other nitrogen-containing xenobiotics. World J. Microbiol. Biotechnol. 2004, 20, 117–135. [Google Scholar] [CrossRef]
- Rieger, P.G.; Meier, H.M.; Gerle, M.; Vogt, U.; Groth, T.; Knackmuss, H.J. Xenobiotics in the environment: Present and future strategies to obviate the problem of biological persistence. J. Biotechnol. 2002, 94, 101–123. [Google Scholar] [CrossRef]
- Schrijver, A.D.; Mot, R.D. Degradation of Pesticides by Actinomycetes. Crit. Rev. Microbiol. 1999, 25, 85–119. [Google Scholar] [CrossRef]
- Bae, K.S.; Kim, M.S.; Lee, J.H.; Kang, J.W.; Kim, D.I.; Lee, J.H.; Seong, C.N. Korean indigenous bacterial species with valid names belonging to the phylum Actinobacteria. J. Microbiol. 2016, 54, 789–795. [Google Scholar] [CrossRef]
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Klenk, H.P.; Clément, C.; Ouhdouch, Y.; van Wezel, G.P. Taxonomy, physiology, and natural products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2015, 80, 1–43. [Google Scholar] [CrossRef]
- Zanini, J.; Deodato, W.; Minillo, A.; de Carvalho, S.L.; Pereira, E.; Pinê, J.H.; Sánchez, I.A. Atrazine herbicide removal through a combination of slow sand filtration and granular activated carbon filters. Rev. Téc. Ing. 2014, 37, 125–134. [Google Scholar]
- Aamand, J.; Jørgensen, C.; Arvin, E.; Jensen, B.K. Microbial adaptation to degradation of hydrocarbons in polluted and unpolluted groundwater. J. Contam. Hydrol. 1989, 4, 299–312. [Google Scholar] [CrossRef]
- Vargha, M.; Takáts, Z.; Márialigeti, K. Degradation of atrazine in a laboratory scale model system with Danube river sediment. Water Res. 2005, 39, 1560–1568. [Google Scholar] [CrossRef]
- Ganesh-Kumar, S.; Kalimuthu, K.; Solomon, R.D.J. A novel bacterium that degrades Aroclor-1254 and its bphC gene encodes an extradiol aromatic ring cleavage dioxygenase (EARCD). Water Air Soil Pollut. 2013, 224, 1587. [Google Scholar] [CrossRef]
- Suzuki, D.; Baba, D.; Satheeja Santhi, V.; Solomon, R.D.J.; Katayama, A. Use of a glass bead-containing liquid medium for efficient production of a soil-free culture with polychlorinated biphenyl-dechlorination activity. World J. Microbiol. Biotechnol. 2013, 29, 1461–1471. [Google Scholar] [CrossRef] [PubMed]
- Piutti, S.; Semon, E.; Landry, D.; Hartmann, A.; Dousset, S.; Lichtfouse, E.; Topp, E.; Soulas, G.; Martin-Laurent, F. Isolation and characterisation of Nocardioides sp. SP12, an atrazine-degrading bacterial strain possessing the gene trzN from bulk- and maize rhizosphere soil. FEMS Microbiol. Lett. 2003, 221, 111–117. [Google Scholar] [CrossRef]
- Devers, M.; Azhari, N.E.; Kolic, N.U.; Martin-Laurent, F. Detection and organization of atrazine-degrading genetic potential of seventeen bacterial isolates belonging to divergent taxa indicate a recent common origin of their catabolic functions. FEMS Microbiol. Lett. 2007, 273, 78–86. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).