Abstract
This paper proposes a classification framework for automatic sleep stage detection in both male and female human subjects by analyzing the electroencephalogram (EEG) data of polysomnography (PSG) recorded for three regions of the human brain, i.e., the pre-frontal, central, and occipital lobes. Without considering any artifact removal approach, the residual neural network (ResNet) architecture is used to automatically learn the distinctive features of different sleep stages from the power spectral density (PSD) of the raw EEG data. The residual block of the ResNet learns the intrinsic features of different sleep stages from the EEG data while avoiding the vanishing gradient problem. The proposed approach is validated using the sleep dataset of the Dreams database, which comprises of EEG signals for 20 healthy human subjects, 16 female and 4 male. Our experimental results demonstrate the effectiveness of the ResNet based approach in identifying different sleep stages in both female and male subjects compared to state-of-the-art methods with classification accuracies of 87.8% and 83.7%, respectively.
1. Introduction
Sleep stage classification can be helpful in identifying sleep disorders, such as, snoring, insomnia, sleep apnea, sleep deprivation, narcolepsy, sleep hypoventilation, and teeth grinding, etc. [1,2]. Sleep disorders are important healthcare concerns as they are significant contributors to fatigue and drowsiness, especially among drivers, and cause around 10–15% of the total vehicle accidents involving fatalities each year [3,4]. Therefore, in order to improve road safety and reduce the risk to millions of human lives, it is important that we understand the causes of sleep disorders, which involves, above all, the understanding and identification of different sleep stages [3,5]. Human sleep is of two fundamental types; rapid eye movement sleep (REM) and non-rapid eye movement sleep (NREM). The NREM sleep is believed to occur in three stages, i.e., N1, N2 and N3, where each stage progressively turns into deeper sleep. Among these NREM stages, most of our sleep time is spent in the N2 stage [6], whereas the REM sleep first starts 90 minutes after we fall asleep, and is mostly associated with dreaming. During a full night’s sleep we go through multiple cycles of REM and NREM sleep [6].
Polysomnography is the study of sleep and is usually undertaken to study sleep disorders. It involves the collection of data from different sources using techniques, such as, electrooculogram (EOG), electrocardiogram (ECG), electroencephalogram (EEG), and electromyogram (EMG). Among these, electrical signals recorded using an EEG are believed to provide more information about brain activity and can be collected in a non-invasive manner [7,8,9]. The placement of electrodes for recording the EEG signal is guided either by the benchmark set by the American Academy of Sleep Medicine (AASM) [10,11]; or by the international 10–20 system for the placement of electrodes [10,12]. The EEG signals considered for the purpose of this research were recorded for three different regions of the brain by placing the electrodes at three different lobes, i.e., Fp2-A1 (pre-frontal), Cz2-A1v (central), and O2-A1 (occipital), following the international 10–20 system. The EEG data was recorded separately for both male and female subjects due to biological differences between the two that could result in variations in their brain activities [13,14,15,16].
Many sleep stage classification techniques have been proposed that use different features of the EEG signal for classification [17,18,19]. However, these techniques mostly use single or dual channel EEG data that is recorded from either one or two regions of the brain. Moreover, the choice of the brain lobes for recording EEG data is mostly arbitrary. Several studies have indicated that multi-channel EEG analysis is vital for accurate sleep stage detection [2,11,20]. Hence, in this study, we consider multi-channel EEG signals (Fp2-A1, Cz2-A1, and O2-A1) for sleep stage classification. Furthermore, existing techniques are not very robust in sleep stage identification, mostly due to the non-stationary nature of the EEG signals, which is not easily captured by the extracted features. Hence, instead of crafting our own features of the EEG signal in time, frequency or time-frequency domains [21,22], a more promising approach would be to use an appropriate deep learning model that would automatically learn distinguishing features from the EEG data to uniquely identify each sleep stage [23]. Visual inspection of EEG recordings has also been considered to uncover unusual patterns in the data, however, it has proved to be difficult and time consuming [24].
Methods based on neural networks [25] and graph based similarity [26] have also been considered for sleep state identification. Neural networks, particularly, deep learning models are efficient at learning a hierarchy of distinguishing features due to their layered architectures [27], but they have their own problems. For instance, conventional deep learning algorithms that use the sigmoid activation functions often encounter the vanishing gradient problem during training [28]. Similarly, deeper network architectures mean more tunable parameters and hence a more complex optimization problem to solve. Hence, stacking more layers may not guarantee better performance [29]. Residual neural networks (ResNet) were proposed to address these shortcomings and were found to be more efficient than the general convolutional neural networks (CNN) [30,31]. ResNet uses several identity shortcuts, also known as residual connections, with the convolution architecture to backpropagate the errors through multiple intermediate layers. Thus, a ResNet can effectively train networks, even with 1001 layers [32]. Moreover, ResNet incorporates many techniques for better training of neural networks, such as momentum, batch normalization, regularization, and weight initialization [30,33].
In this study, a ResNet architecture is employed for automatic sleep state classification. Moreover, instead of the raw EEG signal, its Welch power spectral density (PSD) is calculated and then used as input to the ResNet based classifier. The Welch PSD determines the distribution of signal power over frequency [34], which reduces the estimated power spectra at the cost of reduced frequency resolution [35]. Human EEG studies have revealed that aging reduces the power of lower frequency bands, i.e., delta, theta, and sigma, while increasing that of the higher frequency bands, i.e., beta [36,37]. Moreover, it has also been noted that female human subjects have higher PSD than their male counterparts in the delta and theta range [36,37]. Similarly, in [38], Gabryelska et al. validated that higher individual sleep quality is correlated with reduced NREM stage 2 (N2) sigma 2 spectral power in healthy individuals. Moreover, the amount of variance in subjective sleep quality, which can be described through PSD of EEG, was found to be very small. However, as previously discussed, the ResNet architecture can learn these small changes to classify sleep stages.
The main contributions of this work are as follows:
- An automatic sleep stage detection framework is developed for both genders due to inherent biologoical differences between the two that might affect the electrical activities in their brains and consequently the recorded EEG data.
- The proposed framework considers multi-channel EEG data as input that is recorded at different brain lobes to accurately detect different sleep stages.
- A ResNet architecture, with eight identity shortcut connections, is used with the PSD of time domain EEG signals as input, to identify different sleep stages. The performance of the proposed framework is compared with eight different approaches and is found to be better.
2. Related Work
Safri et al. [39] highlighted the necessity of sex-based sleep state identification and analyzed the recorded EEG signals of 29 students by using partial directed coherence (PDC) and power spectrum estimation (PSE) to investigate gender differences in problem-solving skills. Among other things their work indicated differences between the two genders in terms of the functional connectivity between brain regions, i.e., in PDC, and power distribution of EEG waveforms. Similarly, Chellapa et al. [40] established that gender differences in light sensitivity affect intensity perception, attention and sleep in humans. Silva et al. [16] showed the existence of specific gender differences in sleep pattern by examining the recorded polysomnographic findings of participants in a sleep laboratory, thereby, establishing the need for a gender specific classification approach in sleep stage detection.
Several studies analyzed the attributes of the non-stationary EEG signals from different brain lobes. Wu et al. [17] proposed a signal processing based approach that combined empirical intrinsic geometry (EIG) and the synchro squeezing transform (SST) to compute the dynamic attributes of respiratory and EEG signal of central and occipital scalp sources (C3A2, C4A1, O1A2, and O2A1). Gunes et al. [18] proposed a Welch power spectral density based feature analysis with k-means clustering by analyzing EEG from central (C4-A1) scalp source. Friawan et al. [19] proposed a time-frequency domain-based feature analysis using random forests by analyzing the EEG data from central scalp source (C3-A1). Chen et al. [20] established a decision support algorithm with symbolic fusion for sleep state classification by analyzing EEG data from central scalp source (C3-A2 and C4-A1). Abdulla et al. used correlation graphs coupled with an ensemble model to analyze EEG data from the central scalp source (C3-A2) [26], whereas Phan et al. [27] proposed an attention-based recurrent neural network that used the prefrontal channel (Fpz-Cz) EEG data for sleep stage classification. Most of these methods, however, use EEG data from a single channel, which may not prove to be very robust in sleep stage detection.
Several studies highlighted the importance of considering multichannel EEG analysis to improve the classification performances. Jiang et al. [21] proved the relationship between altitudinal evidence entrenched in multi-channel EEG signals and various sleep stages by utilizing minimum Riemannian distance (RD) to covariance centers, which improved the classification performance. Similarly, Krauss et al. [11] demonstrated that spatially spread cortical activity, exhibited by efficient EEG amplitudes across different recording channels (prefrontal F4-M1, central C4-M1, and occipital O2-M1) contains all the related evidence for separating the sleep stages. Besides, Saper et al. [2] demonstrated that in the pre-frontal lobe (Fp2-A1 channel), the higher executive functions (e.g., emotional regulation, reasoning, and problem solving) occur frequently. In contrast, the visual processing functions are seen mostly in the occipital lobe (O2-A1 channel) of brain [2]. In this study, a classification framework based on multichannel EEG signals (Fp2-A1, Cz2-A1, and O2-A1) is considered.
3. Methodology
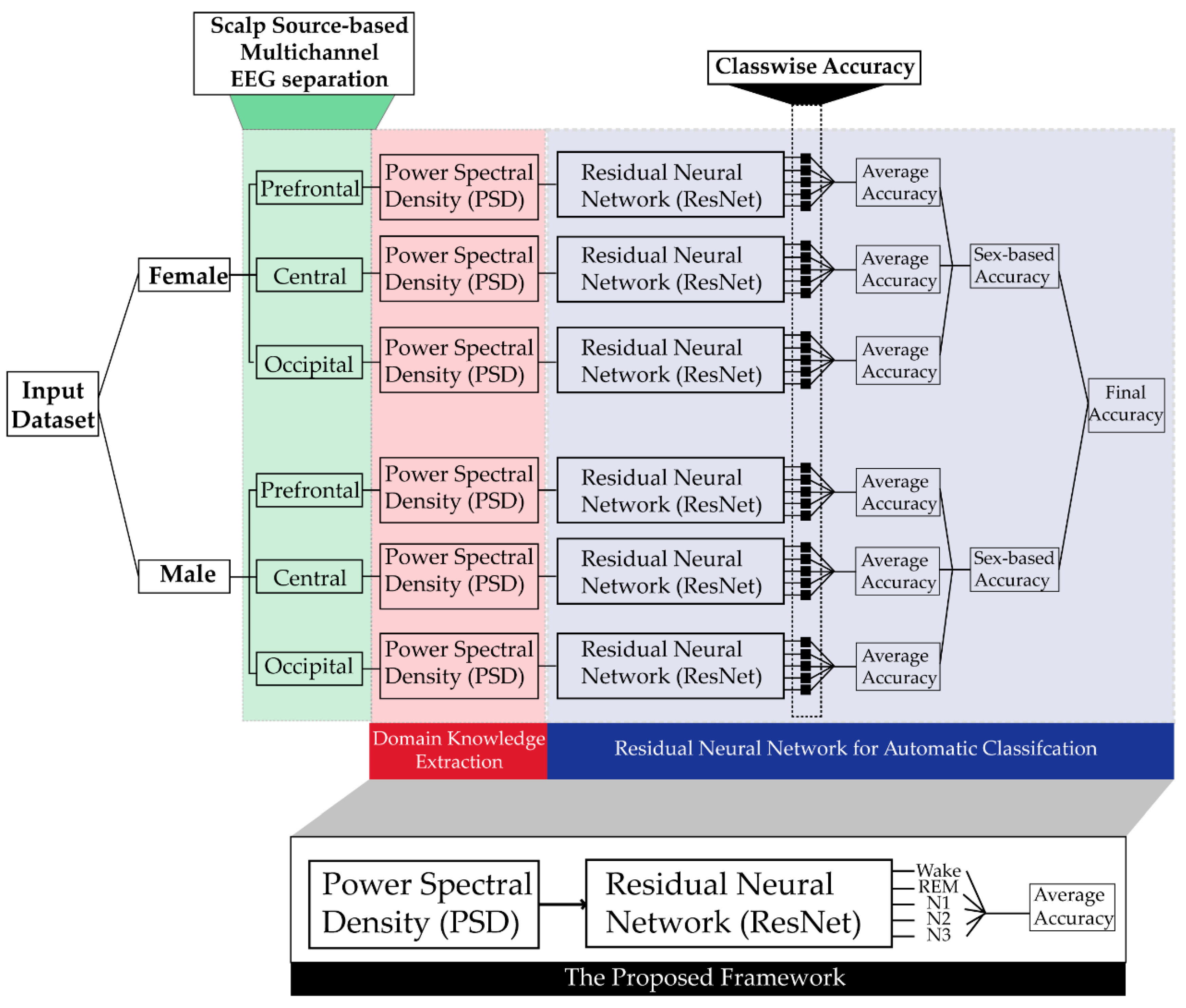
Figure 1 shows the block diagram of the proposed sleep stage classification framework. First, the power spectral density is calculated for individual channels of the EEG data for both the genders. After that, ResNet is trained using the PSD of individual channels as input, to identify different sleep stages based upon input from individual channels that is then combined in subsequent stages. The ResNet is individually trained to identify sleep stages in each gender, and its accuracy is also calculated individually for both genders.
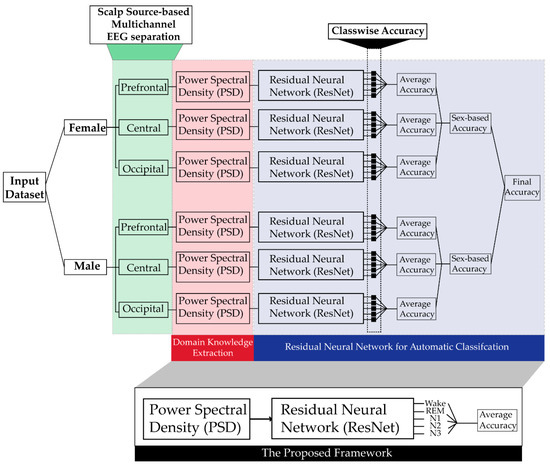
Figure 1.
Block diagram of the proposed method.
3.1. Dataset Description
The proposed approach is validated using a publicly available dataset, i.e., the DREAMS database from the University of MONS–TCTS Laboratory (Stéphanie Devuyst, Thierry Dutoit) and Université Libre de Bruxelles - CHU de Charleroi Sleep Laboratory (Myriam Kerkhofs) [41]. Details of this dataset are given in Table 1. It contains polysomnography (PSG) data or EEG signals of 16 female and 4 male human subjects for a whole night. The data was recorded using a 32 channel polygraph from subjects who were aged between 20 to 65 years. The EEG signals were sampled at a frequeny of 200 Hz. The sleep stages were annotated by experts following analysis of microevents, each of 30-second epoch, using the criteria defined by AASM. As mentioned earlier, data from 3 EEG channels (i.e., Fp2-A1, Cz2-A1, and O2-A1) is used in this study. Moreover, the dataset appears to be a little imbalanced in terms of gender. However, this bias in data is removed by grouping the subjects based upon their gender, i.e., the proposed methodology considers sleep stage detection individually for both genders, hence, the class imbalance in the dataset does not affect the classifier’s performance. In addition, the same number of samples are selected from different sleep stages to create a well-balanced dataset.

Table 1.
Details of considered dataset.
To the best of the authors’ knowledge, very few studies that use data driven techniques have utilized this dataset. Therefore, to establish the robustness of the proposed approach, it is compared with eight different techniques while using the same dataset and experimental setup, i.e., (1) Raw EEG + ResNet, (2) PSD + CNN (5 layers [42]), (3) PSD + CNN (10 layers), (4) PSD + CNN (14 layers), (5) PSD + CNN (18 layers), (6) FE + Random Forest [43] (RF), (7) fast Fourier transform (FFT) + multilayered perceptron (MPC) [44], and (8) PSD + MPC.
3.2. Domain Knowledge Extraction by Power Spectral Density
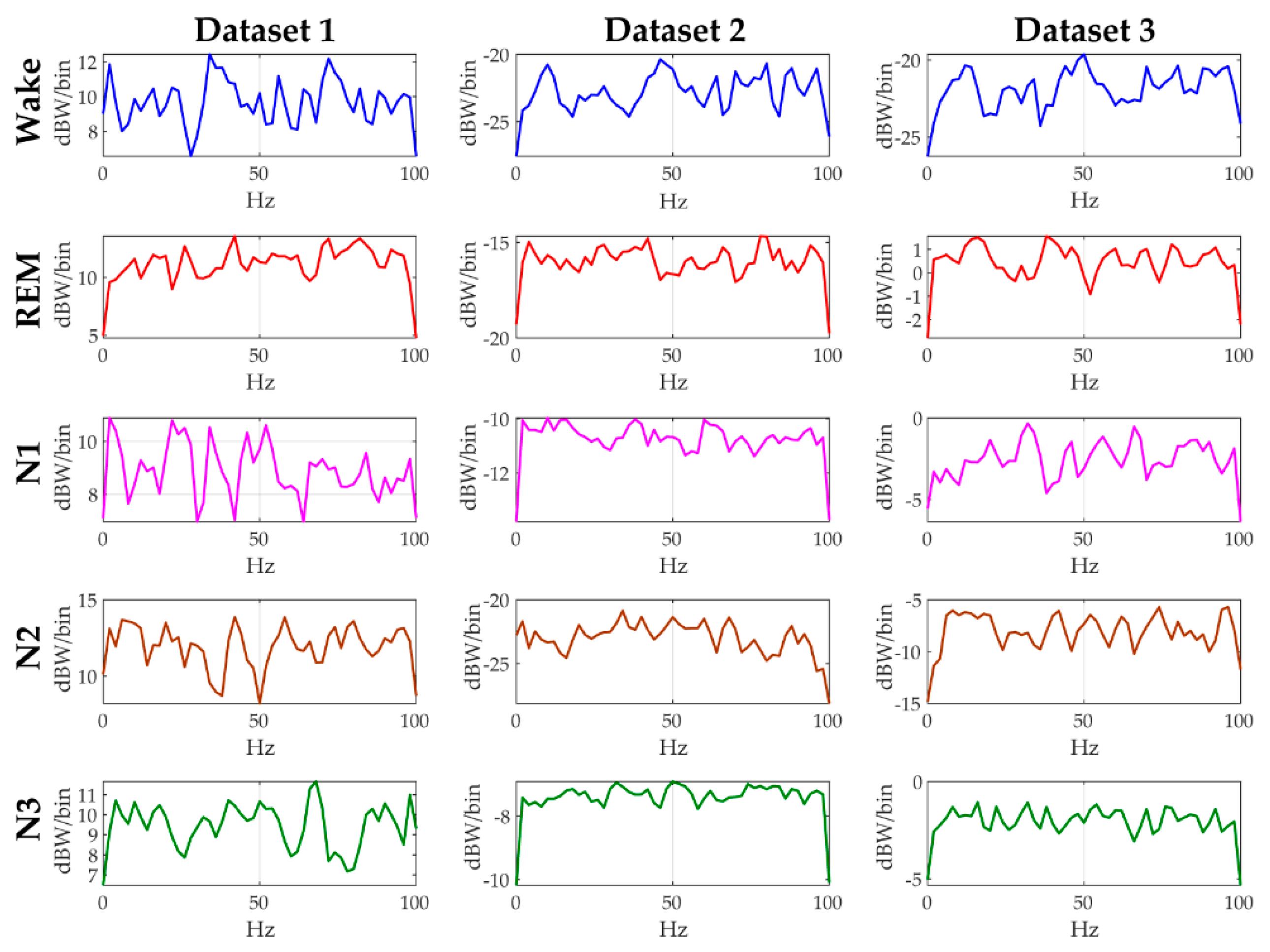
Sleep signals are continuous time series data with fluctuations in amplitude and behavior of different frequencies that changes over time for every sleep stage. The sheer volume of the highly fluctuating raw EEG data makes it challenging for machine learning algorithms to find a meaningful pattern that can be helpful in identifying different sleep stages. Hence, we calculate the Welch PSD of the raw EEG data to improve our chances of finding out a meaningful pattern that can be helpful in identifying different sleep stages [45,46,47,48]. The Welch PSD calculates the distribution of energy across different frequencies of the EEG signal. In Figure 2, the computed Welch PSD is plotted for the five different sleep stages of a female subject to show the differences among the PSD of different sleep stages. For calculating the PSD, the 30 s epoch is considered as an input. After that, DFT of 50% overlapping segment of nfft (number of points in DFT) is used (sampling frequency = 200 Hz, nfft = 100) to calculate the spectrum with DFT averaging. Therefore, the change in the amplitude is observed by converting the amplitude response from dBW/Hz to dBW/bin. For each dataset, the computed PSD response of each sleep state is different from the others, hence necessitating the use of three channel EEG data for sleep state identification.
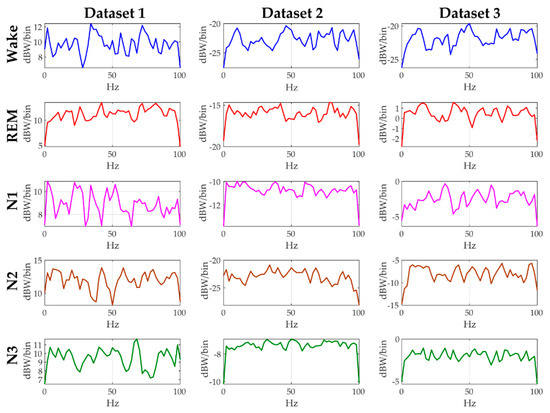
Figure 2.
Power spectral density (PSD) curves of all the sleep stages of a single female subject but from different datasets.
3.3. Residual Neural Network
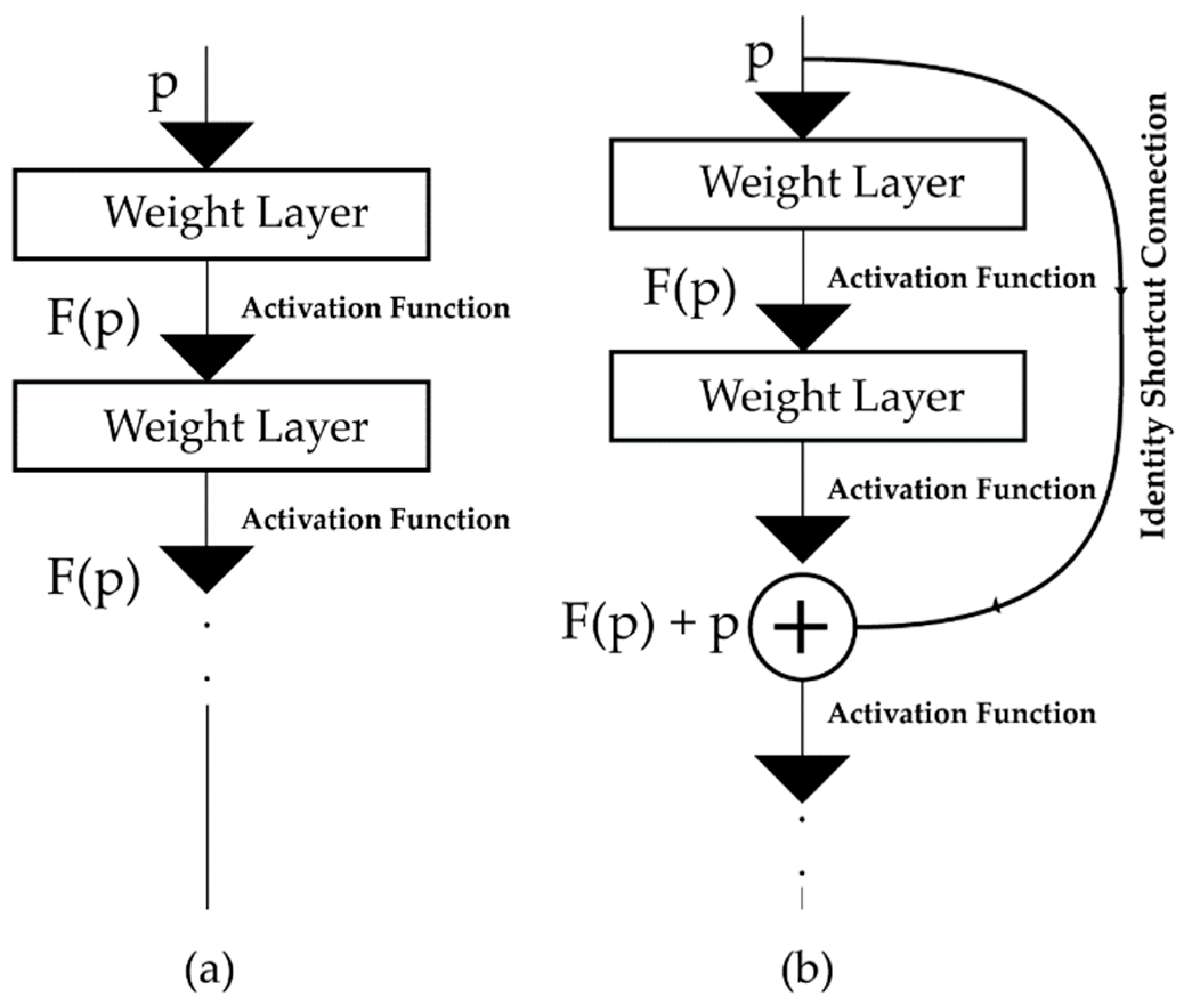
ResNet has emerged as a revolutionary idea in deep learning [28,29]. The main difference between a typical and a residual neural network is the residual connection, i.e., identity or shortcut connection, which makes a significant difference in architecture and performance [49]. Unlike traditional neural networks, where each layer feeds into the next layer (Figure 3a), in a residual neural network, each layer feeds not just into the next layer but also into the layers which are a few hops away [30] (Figure 3b). Thus, it helps to propagate larger gradients to the initial layers through backpropagation, thereby solving the vanishing gradient problem and enabling the training of deeper networks [30,50].
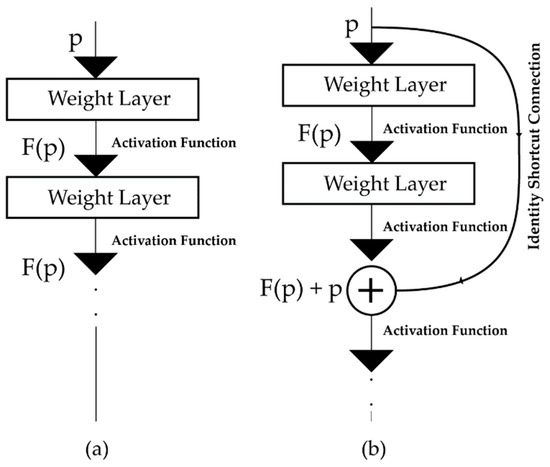
Figure 3.
The connection mechanism of the feed-forward layers for neural architectures, i.e., (a) traditional neural networks, and (b) the residual neural network (ResNet) architecture.
In Figure 3b, is the input of a neural network block, where the network wants to learn the true distribution of the input, denoted as . If the difference (residual) between the input and input distribution is , then
In this work, the calculated PSD is considered as the input to the ResNet. The ResNet architecture in the proposed framework is based on ResNet–18 [30,51,52,53].
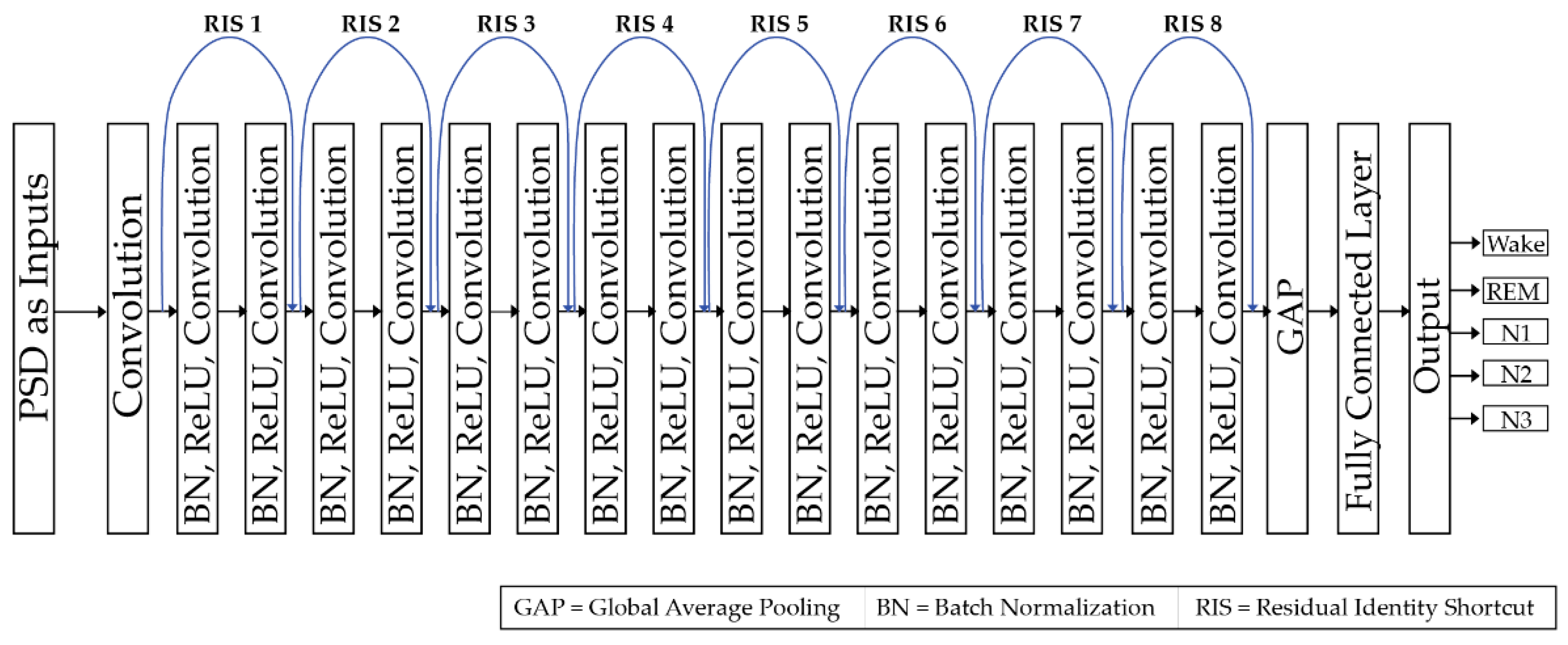
Like the standard CNN [49], this ResNet architecture contains convolution layers, fully connected layers, activation functions, batch normalizations (BN) [54], and a feed forward architecture [31,49]. The weights and biases in the convolutional layers are optimized through the backpropagation of errors during the training, whereas, non-linear transformation is achieved by using the rectified linear unit (ReLU) activation function [55], given by Equation (2). For minimizing the loss function, Adamax optimizer [56] is used. The considered ResNet architecture is shown in Figure 4.
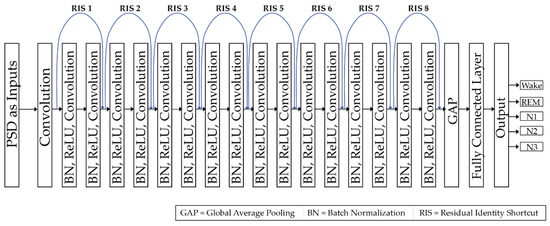
Figure 4.
Block diagram of the proposed architecture.
There are 2 main differences between the original ResNet-18 architecture and the one used in this work, i.e., (a) BN is adopted to deal with internal covariate shift problem mostly due to the non-stationary nature of the PSD input data, and (b) usage of total 8 identity connections throughout the network architecture rather than usage of skip connection [53]. These 8 identity connections allow the gradients to promptly roll through the network architecture, without passing through the non-liner activation functions. This suggests that the proposed deep network model should not create a training error higher than its shallower counterparts. Unfortunately, there is no rule of thumb that either helps to select the best number of identity connections or allows to tell the exact number of layers in the deeper network. It always depends on the complexity of the experimental dataset. The dataset utilized in this study is difficult to handle using traditional deep neural networks because of the nature of the PSD patterns obtained from the EEG signals. Therefore, a deeper network architecture, ResNet, is considered [51,52,53].
3.4. Data Preparation for Classification and Parameters for Performance Measurement
In the dataset used for this study, each sample from individual sleep stage consists of a 30 s epoch length. However, there is a significant imbalance in the data in terms of the number of samples for different sleep stages of individual subjects. To balance the dataset prior to its use by the ResNet, 70 data samples from each sleep stage are randomly selected. Thus, a total of 350 samples are considered for each healthy subject (each class comprises 70 data samples, and in total 5 signal classes were considered in this study). To compute the classification performance of ResNet, K-Fold cross validation (CV) [57] is used. It should be noted that the choice of K, in K-Fold CV is usually arbitrary. To lower the variance of the CV results, it is recommended to repeat/iterate CV procedure with several new random splits. Therefore, in this experiment, the number of K in the CV is 4. For executing the 4-Fold CV, all the samples from the individual datasets are divided in the ratio 75:25 for training and testing, respectively. For example, dataset 1 consists of 5600 samples (350 samples from each of the 16 female subjects), of which 4200 samples are used for training whereas the remaining 1,400 are used for testing. The details of datasets used for training and testing the ResNet are given in Table 2.

Table 2.
Details of datasets used for training and testing the ResNet.
The performance of the proposed framework is evaluated using several parameters, i.e., (a) F1 score (F1) [58], (b) accuracy score (AS), and (c) confusion matrix [59]. The F1 score and AS can be calculated using Equations (3), and (4), respectively.
In Equations (3) and (4), the terms TP, TN, FN, and FP represent the number of true positives, true negetives, false negatives, and false positives, respectively. For measuring the class-wise performance, F1 score is considered. The classification accuracy for each gender is determined by calculating the mean AS. Similarly, the overall accuracy can be determined by calculating the mean accuracy for both genders. During training, the loss function of the proposed deep learning framework is monitored to avoid both overfitting and underfitting [60]. The details of the performance analysis are given in Table 3.

Table 3.
Analysis of the classification accuracy.
4. Results and Discussion
4.1. Performance Analysis of Residual Neural Network
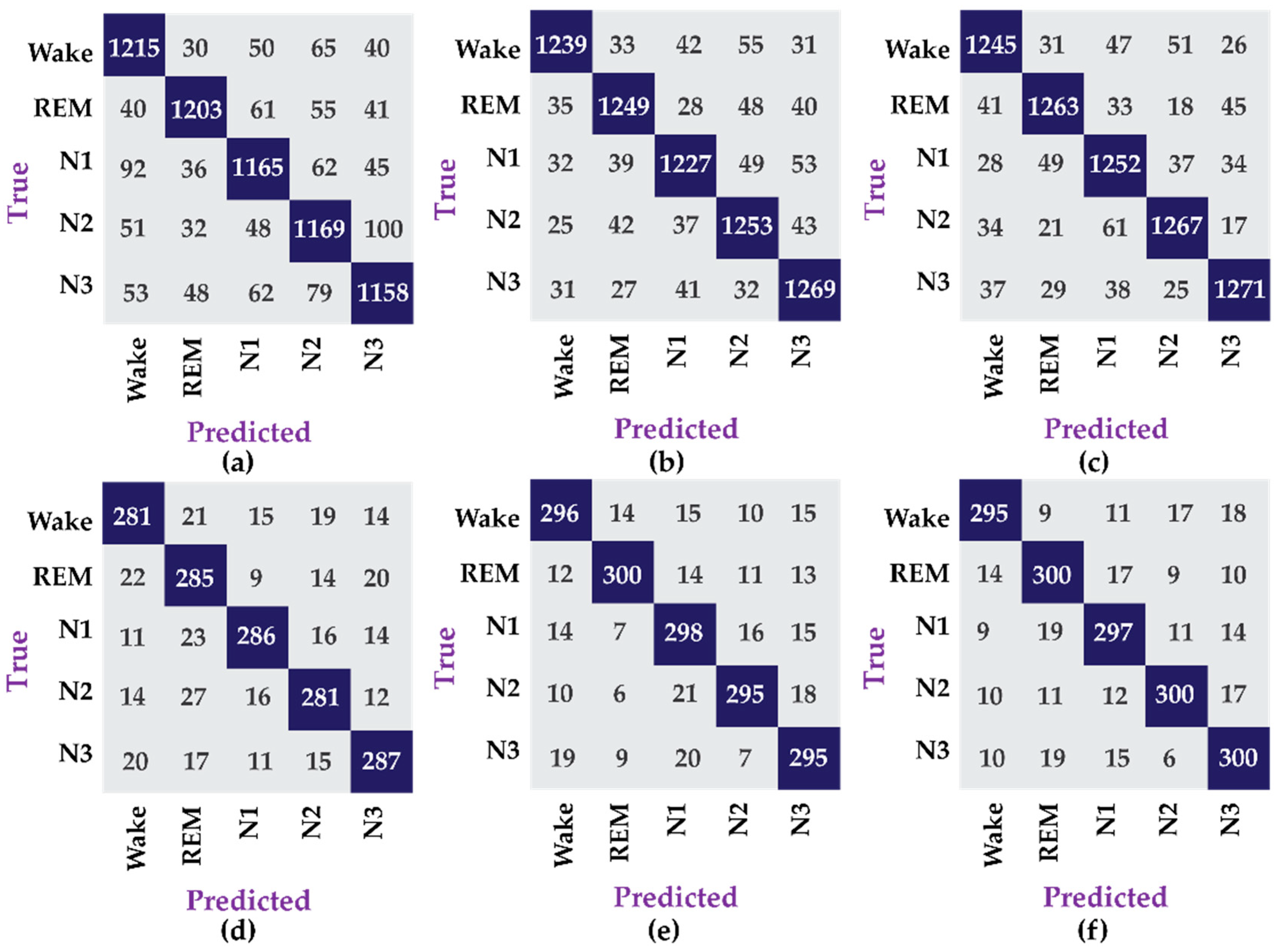
The results in Table 3 show that the data for female subjects yields better performance, 4.1% higher, compared to the data for male subjects, which is due to the fact that the amount of data available for the former is four times more than the later. The amount of data is critical for the construction of good machine learning models particularly deep learning models, which have significantly more parameters to tune. Similarly, the confusion matrices for this experiment are shown in Figure 5.
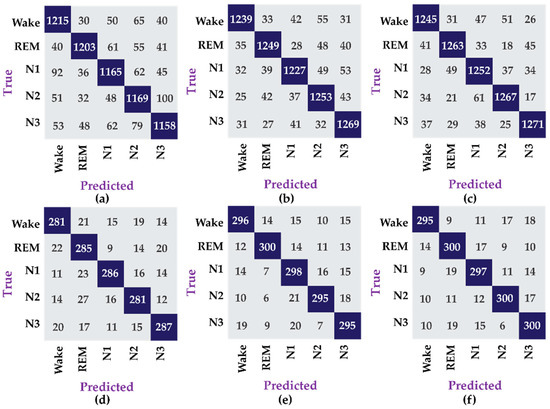
Figure 5.
Confusion matrix for different datasets, i.e., (a) dataset 1, (b) dataset 2, (c) dataset 3, (d) dataset 4, (e) dataset 5, and (f) dataset 6.
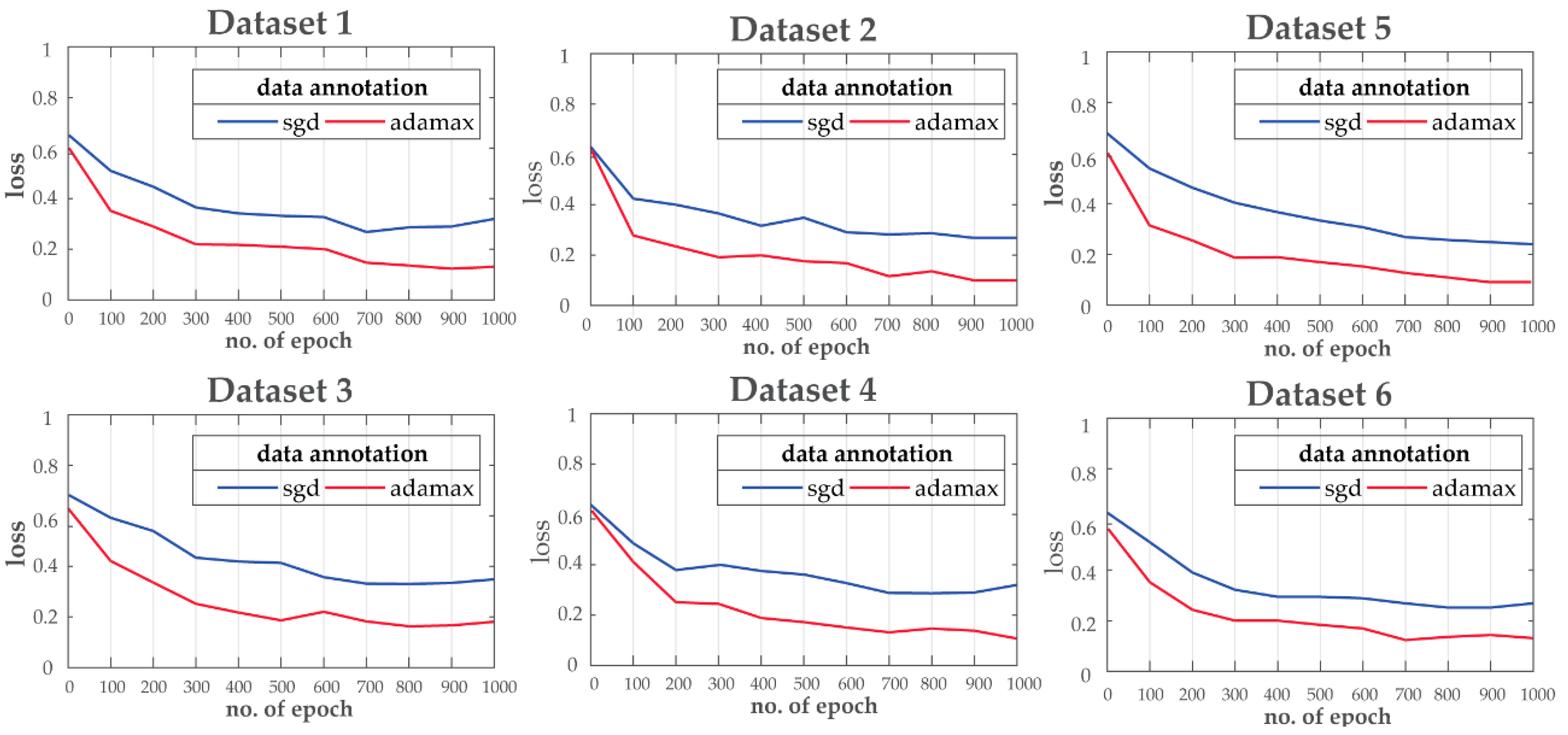
For the purpose of this experiment, the ResNet is trained for 1000 epochs with a 75/25 train/validation ratio to tackle the overfitting-underfitting problem. Moreover, during training, two different optimization techniques: i.e., stochastic gradient descent (SGD) and Adamax are considered for loss function optimization. Adamax shows better convergence (learning rate) for all the datasets compared to SGD as shown in Figure 6.
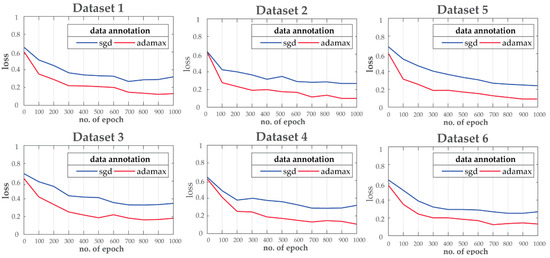
Figure 6.
Training loss curve comparisons for different datasets.
4.2. Comparisons and Discussion
The proposed approach is compared with the following eight techniques using the same datasets:
(1) Raw EEG + ResNet: this approach uses the raw multi-channel EEG signals as input to the ResNet architecture used in the proposed framework. Comparison with this technique highlights the benefit of using the PSD of the EEG signals over the raw EEG signals as input.
(2) PSD + CNN (5 layers): this approach uses the same input data, i.e., PSD of raw EEG signals, but uses a 5-layer CNN instead of a ResNet. Comparison with this technique highlights the benefit of using ResNet compared to CNN.
(3) PSD + CNN (10 layers): this approach uses the same input data, i.e., PSD of raw EEG signals, but uses a 10-layer CNN instead of a ResNet. Comparison with this technique highlights the benefit of using ResNet compared to a CNN with 10 layers.
(4) PSD + CNN (14 layers): this approach uses the same input data, i.e., PSD of raw EEG signals, but uses a 10-layer CNN instead of a ResNet. Comparison with this technique and the two techniques prior to it highlights the benefit of using ResNet compared to CNN with increasing number of layers.
(5) PSD + CNN (18 layers): this approach uses the same input data, i.e., PSD of raw EEG signals, but uses an 18-layer CNN instead of a ResNet. The number of layers in the CNN is the same as the number of layers in the ResNet in the proposed framework. Nevertheless, the performance of ResNet is better in comparison.
(6) FE + RF: this approach uses time-frequency based features extraction (FE) and a random forest (RF) classifier [41]. Comparison with this approach highlights the significance of both the input data and ResNet architecture used in the proposed framework.
(7) FFT + MPC: this approach uses fast Fourier transform (FFT) based feature analysis and a multilayered perceptron (MPC) based classifier [42] to highlight the importance of the deeper residual network architecture.
(8) PSD + MPC: this approach uses the PSD of EEG signals as input to a MPC based classifier to highlight the importance of the deeper residual network architecture.
Comparison of all these approaches with the proposed framework is given in Table 4.

Table 4.
Accuracy comparison of different methods.
The results in Table 4 clearly show that the proposed approach is significantly more effective than all the other approaches in correctly identifying sleep stages in both the genders. The “Improvement” column in Table 4 shows the percentage improvement in absolute terms that the proposed method has over the other techniques. The comparison results show that the proposed framework (PSD + ResNet) yields an average performance improvement of 3.8–40.6% and 1.3–37.9% for female and male subjects, respectively.
The results in Table 3 show that the performance of raw EEG + ResNet is inferior to the proposed PSD + ResNet based framework, which indicates that the ResNet architecture is more efficient in learning the smaller changes of the subjective sleep states from the PSD compared to the raw EEG. Achieving the same performance with the raw EEG as input would require a deeper ResNet architecture with more identity shortcut connections. However, as the input length of raw EEG is far greater than the PSD, it would increase the training time for the ResNet. Moreover, approaches based on deep learning do not require artifact removal from the raw data, since these approaches are very good at automatically learning small changes in the data distribution [30,31,49].
5. Conclusions
In this study, an automatic sleep state detection framework is presented for both male and female human subjects. The proposed framework uses the PSD of EEG signals collected from three important regions of the brain, i.e., the pre-frontal, central, and occipital, as input to a ResNet. The ResNet architecture automatically extracts the intrinsic information for each sleep stage to classify it correctly. The proposed framework was tested on the publicly available Dreams dataset. It achieved accuracies of 87.8% and 83.7% for female and male subjects, respectively. The proposed framework was compared with several state-of-the-art methods to justify the choice of a ResNet based classifier and PSD based input data, and it yielded significant performance improvements, i.e., 3.8–40.6% and 1.3–37.9% for female and male subjects, respectively. The dataset used for validation of the proposed framework is imbalanced in terms of gender, which inevitably affected the experimental results. The difference in performance is understandable as in machine learning as well as in deep learning, more data often translates into a better model. Nevertheless, in future, a more extensive and balanced dataset would be considered to validate the findings of this study. Furthermore, in addition to PSD, other time-frequency based analysis techniques will be explored to capture all the variations in the non-stationary EEG signals.
Author Contributions
All the authors contributed equally to the conception of the idea, implementing, and analyzing the experimental results, and writing the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by an Electronics and Telecommunications Research Institute (ETRI) grant funded by the Korean government. [20ZS1200, Research on original technology of human-centered autonomous intelligence system/20AS1100, the development of smart HSE and digital cockpit systems based on ICT convergence for enhanced major industry].
Conflicts of Interest
The authors declare no conflict of interest.
References
- Morin, C.M.; Espie, C.A. The Oxford Handbook of Sleep and Sleep Disorders; Oxford University Press: Oxford, UK, 2011; ISBN 0199704422. [Google Scholar]
- Saper, C.B.; Fuller, P.M.; Pedersen, N.P.; Lu, J.; Scammell, T.E. Sleep state switching. Neuron 2010, 68, 1023–1042. [Google Scholar] [CrossRef] [PubMed]
- Supriya, S.; Siuly, S.; Wang, H.; Zhang, Y. EEG Sleep Stages Analysis and Classification Based on Weighed Complex Network Features. IEEE Trans. Emerg. Top. Comput. Intell. 2018, PP, 1–11. [Google Scholar] [CrossRef]
- Altevogt, B.M.; Colten, H.R. Sleep Disorders and Sleep Deprivation: An Unmet Public Health Problem; National Academies Press: Washington, DC, USA, 2006; ISBN 0309101115. [Google Scholar]
- Ferrie, J.E.; Kumari, M.; Salo, P.; Singh-Manoux, A.; Kivimäki, M. Sleep epidemiology—A rapidly growing field. Int. J. Epidemiol. 2011, 40, 1431–1437. [Google Scholar] [CrossRef]
- Malik, J.; Lo, Y.-L.; Wu, H. Sleep-wake classification via quantifying heart rate variability by convolutional neural network. Physiol. Meas. 2018, 39, 85004. [Google Scholar] [CrossRef] [PubMed]
- Von Rosenberg, W.; Chanwimalueang, T.; Goverdovsky, V.; Looney, D.; Sharp, D.; Mandic, D.P. Smart helmet: Wearable multichannel ECG and EEG. IEEE J. Transl. Eng. Heal. Med. 2016, 4, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Siuly, S.; Zhang, Y. Medical big data: Neurological diseases diagnosis through medical data analysis. Data Sci. Eng. 2016, 1, 54–64. [Google Scholar] [CrossRef]
- Kabir, E.; Siuly, S.; Cao, J.; Wang, H. A computer aided analysis scheme for detecting epileptic seizure from EEG data. Int. J. Comput. Intell. Syst. 2018, 11, 663–671. [Google Scholar] [CrossRef]
- Berry, R.B. The AASM Manual for the Scoring of Sleep and Associated Events: Rules, Terminology and Technical Specifications; American Academy of Sleep Medicine: Darien, IL, USA, 2018. [Google Scholar]
- Krauss, P.; Schilling, A.; Bauer, J.; Tziridis, K.; Metzner, C.; Schulze, H.; Traxdorf, M. Analysis of Multichannel EEG Patterns During Human Sleep: A Novel Approach. Front. Hum. Neurosci. 2018, 12, 121. [Google Scholar] [CrossRef]
- Binnie, C.D.; Dekker, E.; Smit, A.; Van der Linden, G. Practical considerations in the positioning of EEG electrodes. Electroencephalogr. Clin. Neurophysiol. 1982, 53, 453–458. [Google Scholar] [CrossRef]
- Ujma, P.P.; Konrad, B.N.; Simor, P.; Gombos, F.; Körmendi, J.; Steiger, A.; Dresler, M.; Bódizs, R. Sleep EEG functional connectivity varies with age and sex, but not general intelligence. Neurobiol. Aging 2019, 78, 87–97. [Google Scholar] [CrossRef]
- Tomescu, M.I.; Rihs, T.A.; Rochas, V.; Hardmeier, M.; Britz, J.; Allali, G.; Fuhr, P.; Eliez, S.; Michel, C.M. From swing to cane: Sex differences of EEG resting-state temporal patterns during maturation and aging. Dev. Cogn. Neurosci. 2018, 31, 58–66. [Google Scholar] [CrossRef]
- Ferrara, M.; Bottasso, A.; Tempesta, D.; Carrieri, M.; De Gennaro, L.; Ponti, G. Gender differences in sleep deprivation effects on risk and inequality aversion: Evidence from an economic experiment. PLoS ONE 2015, 10, e0120029. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.; Andersen, M.L.; De Mello, M.T.; Bittencourt, L.R.A.; Peruzzo, D.; Tufik, S. Gender and age differences in polysomnography findings and sleep complaints of patients referred to a sleep laboratory. Braz. J. Med. Biol. Res. 2008, 41, 1067–1075. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Talmon, R.; Lo, Y.-L. Assess sleep stage by modern signal processing techniques. IEEE Trans. Biomed. Eng. 2014, 62, 1159–1168. [Google Scholar] [CrossRef] [PubMed]
- Güneş, S.; Polat, K.; Yosunkaya, Ş. Efficient sleep stage recognition system based on EEG signal using k-means clustering based feature weighting. Expert Syst. Appl. 2010, 37, 7922–7928. [Google Scholar] [CrossRef]
- Fraiwan, L.; Lweesy, K.; Khasawneh, N.; Wenz, H.; Dickhaus, H. Automated sleep stage identification system based on time–frequency analysis of a single EEG channel and random forest classifier. Comput. Methods Programs Biomed. 2012, 108, 10–19. [Google Scholar] [CrossRef]
- Chen, C.; Ugon, A.; Zhang, X.; Amara, A.; Garda, P.; Ganascia, J.-G.; Philippe, C.; Pinna, A. Personalized sleep staging system using evolutionary algorithm and symbolic fusion. In Proceedings of the 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 16–20 August 2016; IEEE: Washington, DC, USA, 2016; pp. 2266–2269. [Google Scholar]
- Jiang, D.; Ma, Y.; Wang, Y. Sleep stage classification using covariance features of multi-channel physiological signals on Riemannian manifolds. Comput. Methods Programs Biomed. 2019, 178, 19–30. [Google Scholar] [CrossRef]
- Sermanet, P.; Chintala, S.; LeCun, Y. Convolutional neural networks applied to house numbers digit classification. arXiv 2012, arXiv:1204.3968. [Google Scholar]
- Vrbancic, G.; Podgorelec, V. Automatic classification of motor impairment neural disorders from EEG signals using deep convolutional neural networks. Elektron. Elektrotechnika 2018, 24, 3–7. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, Y. Automatic sleep stage classification of single-channel EEG by using complex-valued convolutional neural network. Biomed. Eng. Tech. 2018, 63, 177–190. [Google Scholar] [CrossRef]
- Ebrahimi, F.; Mikaeili, M.; Estrada, E.; Nazeran, H. Automatic sleep stage classification based on EEG signals by using neural networks and wavelet packet coefficients. In Proceedings of the 2008 30th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Vancouver, BC, Canada, 20–25 August 2008; IEEE: Washington, DC, USA, 2008; pp. 1151–1154. [Google Scholar]
- Abdulla, S.; Diykh, M.; Laft, R.L.; Saleh, K.; Deo, R.C. Sleep EEG signal analysis based on correlation graph similarity coupled with an ensemble extreme machine learning algorithm. Expert Syst. Appl. 2019, 138, 112790. [Google Scholar] [CrossRef]
- Phan, H.; Andreotti, F.; Cooray, N.; Chen, O.Y.; Vos, M. De Automatic Sleep Stage Classification Using Single-Channel EEG: Learning Sequential Features with Attention-Based Recurrent Neural Networks. Proc. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. EMBS 2018, 2018, 1452–1455. [Google Scholar]
- Zhao, M.; Kang, M.; Tang, B.; Pecht, M. Deep Residual Networks with Dynamically Weighted Wavelet Coefficients for Fault Diagnosis of Planetary Gearboxes. IEEE Trans. Ind. Electron. 2018, 65, 4290–4300. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Delving deep into rectifiers: Surpassing human-level performance on imagenet classification. In Proceedings of the IEEE International Conference on Computer Vision 2015, Santiago, Chile, 7–13 December 2015; pp. 1026–1034. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar]
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2323. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. In Proceedings of the Advances in Neural Information Processing Systems, Lake Tahoe, NV, USA, 3–6 December 2012; pp. 1097–1105. [Google Scholar]
- Zhao, M.; Kang, M.; Tang, B.; Pecht, M. Multiple Wavelet Coefficients Fusion in Deep Residual Networks for Fault Diagnosis. IEEE Trans. Ind. Electron. 2019, 66, 4696–4706. [Google Scholar] [CrossRef]
- Rajak, B.L.; Gupta, M.; Bhatia, D.; Mukherjee, A.; Paul, S.; Sinha, T.K. Power Spectral Analysis of EEG as a Potential Marker in the Diagnosis of Spastic Cerebral Palsy Cases. Int. J. Biomed. Eng. Sci. 2016, 3, 23–29. [Google Scholar] [CrossRef]
- Unde, S.A.; Shriram, R. PSD Based Coherence Analysis of EEG Signals for Stroop Task. Int. J. Comput. Appl. 2014, 95, 1–5. [Google Scholar] [CrossRef]
- Carrier, J.; Land, S.; Buysse, D.J.; Kupfer, D.J.; Monk, T.H. The effects of age and gender on sleep EEG power spectral density in the middle years of life (ages 20–60 years old). Psychophysiology 2001, 38, 232–242. [Google Scholar] [CrossRef]
- Fernandez, L.M.J.; Luthi, A. Sleep Spindles: Mechanisms and Functions. Physiol. Rev. 2019, 100, 805–868. [Google Scholar] [CrossRef]
- Gabryelska, A.; Feige, B.; Riemann, D.; Spiegelhalder, K.; Johann, A.; Białasiewicz, P.; Hertenstein, E. Can spectral power predict subjective sleep quality in healthy individuals? J. Sleep Res. 2019, 28, e12848. [Google Scholar] [CrossRef]
- The DREAMS Subjects Database. Available online: https://zenodo.org/record/2650142#.Xy-n5igzYkl (accessed on 16 September 2020).
- LeCun, Y.; Jackel, L.D.; Bottou, L.; Cortes, C.; Denker, J.S.; Drucker, H.; Guyon, I.; Muller, U.A.; Sackinger, E.; Simard, P. Learning algorithms for classification: A comparison on handwritten digit recognition. Neural Networks Stat. Mech. Perspect. 1995, 261, 276. [Google Scholar]
- Bi, S.; Liao, Q.; Lu, Z. An Approach of Sleep Stage Classification Based on Time-frequency Analysis and Random Forest on a Single Channel. ACAAI 2018, 155, 209–213. [Google Scholar]
- Belakhdar, I.; Kaaniche, W.; Djmel, R.; Ouni, B. A comparison between ANN and SVM classifier for drowsiness detection based on single EEG channel. In Proceedings of the 2nd International Conference on Advanced Technologies for Signal and Image Processing (ATSIP 2016), Monastir, Tunisia, 21–23 March 2016; IEEE: Washington, DC, USA, 2016; pp. 443–446. [Google Scholar]
- Safri, N.M.; Sha’ameri, A.Z.; Samah, N.A.; Daliman, S. Resolving Gender Difference in Problem Solving Based On the Analysis of Electroencephalogram (EEG) Signals. Int. J. Integr. Eng. 2018, 10, 10. [Google Scholar]
- Chellappa, S.L.; Steiner, R.; Oelhafen, P.; Cajochen, C. Sex differences in light sensitivity impact on brightness perception, vigilant attention and sleep in humans. Sci. Rep. 2017, 7, 14215. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, M.S. Periodogram analysis and continuous spectra. Biometrika 1950, 37, 1–16. [Google Scholar] [CrossRef]
- Bartlett, M.S.; Medhi, J. On the efficiency of procedures for smoothing periodograms from time series with continuous spectra. Biometrika 1955, 42, 143–150. [Google Scholar] [CrossRef]
- Christensen, J.A.E.; Munk, E.G.S.; Peppard, P.E.; Young, T.; Mignot, E.; Sorensen, H.B.D.; Jennum, P. The diagnostic value of power spectra analysis of the sleep electroencephalography in narcoleptic patients. Sleep Med. 2015, 16, 1516–1527. [Google Scholar] [CrossRef] [PubMed]
- Rahi, P.K.; Mehra, R. Analysis of power spectrum estimation using welch method for various window techniques. Int. J. Emerg. Technol. Eng. 2014, 2, 106–109. [Google Scholar]
- Lecun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Identity mappings in deep residual networks. In Proceedings of the European Conference on Computer Vision, Amsterdam, The Netherlands, 11–14 October 2016; Springer: Cham, Switzerland, 2016; pp. 630–645. [Google Scholar]
- Lin, X.; Zhao, C.; Pan, W. Towards accurate binary convolutional neural network. In Proceedings of the Advances in Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; pp. 345–353. [Google Scholar]
- Pakhomov, D.; Premachandran, V.; Allan, M.; Azizian, M.; Navab, N. Deep residual learning for instrument segmentation in robotic surgery. In Proceedings of the International Workshop on Machine Learning in Medical Imaging; Springer: Cham, Switzerland, 2019; pp. 566–573. [Google Scholar]
- Khan, R.U.; Zhang, X.; Kumar, R.; Aboagye, E.O. Evaluating the performance of resnet model based on image recognition. In Proceedings of the 2018 International Conference on Computing and Artificial Intelligence, Chengdu, China, 12–14 March 2018; ACM: New York, NY, USA, 2018; pp. 86–90. [Google Scholar]
- Zhou, P.; Austin, J. Learning criteria for training neural network classifiers. Neural Comput. Appl. 1998, 7, 334–342. [Google Scholar] [CrossRef]
- Glorot, X.; Bordes, A.; Bengio, Y. Deep sparse rectifier neural networks. In Proceedings of the Fourteenth International Conference on Artificial Intelligence and Statistics, Fort Lauderdale, FL, USA, 11–13 April 2011; pp. 315–323. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Browne, M.W. Cross-validation methods. J. Math. Psychol. 2000, 44, 108–132. [Google Scholar] [CrossRef]
- Luque, A.; Carrasco, A.; Martín, A.; de las Heras, A. The impact of class imbalance in classification performance metrics based on the binary confusion matrix. Pattern Recognit. 2019, 91, 216–231. [Google Scholar] [CrossRef]
- Goutte, C.; Gaussier, E. A probabilistic interpretation of precision, recall and F-score, with implication for evaluation. In Proceedings of the European Conference on Information Retrieval, Santiago de Compostela, Spain, 21–23 March 2005; Springer: Cham, Switzerland, 2005; pp. 345–359. [Google Scholar]
- Goodfellow, I.; Bengio, Y.; Courville, A. Machine learning basics. Deep Learn. 2016, 1, 98–164. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).