1. Introduction
In many real-world medical domain problems, the datasets collected for data mining purposes are usually incomplete, containing missing (attribute) values or missing data, such as pulmonary embolism data [
1], DNA microarray data [
2], metabolomics data [
3], cardiovascular disease data [
4], lung disease data [
5], food composition data [
6], traffic data [
7], and other medical data [
8].
Many data mining and machine learning algorithms used in the data mining process are not able to effectively analyze incomplete datasets. In addition, directly using incomplete datasets for the purpose of data analysis can have a significant effect on the final conclusions that are drawn from the data [
9].
There are a number of different techniques that can be used to deal with missing values, such as case deletion, mean substitution, and model-based imputation, to name a few [
10,
11,
12]. Among them, the simplest solution is based on case deletion (or listwise deletion), in which data containing missing values are deleted. However, it is problematic when missing data are not random or the missing rate for the whole dataset is larger than a certain value, for example 10% [
11,
13].
Model-based imputation methods using machine learning techniques have been shown to outperform many other statistical techniques [
14,
15,
16,
17,
18,
19]. In general, these types of model-based imputation methods are based on machine learning techniques and involve training using a set of complete data to produce estimations to replace the missing values in an incomplete dataset.
However, since a collected (incomplete) dataset must contain a number of features (i.e., input variables) to represent the data, it is likely that some of the features will not be representative, which can affect the discriminatory power of the data mining algorithms. In other words, redundant and irrelevant features or unwanted features from the collected dataset must be filtered out; otherwise the mining performance will be affected. This situation could be even worse when ultra-high or hyperdimensional datasets containing a very large number of features are used, which is called the curse of dimensionality [
20].
For the purpose of missing value imputation, performing feature selection over the observed data to filter out unrepresentative features could make the imputation process more efficient, since some of the missing features, which may be regarded as unrepresentative, are not required for imputation. Moreover, feature selection is able to make the imputation model trained by the lower dimensional observed data provide better estimations for the rest of the missing features.
In literature, several studies have focused on this issue [
21,
22]. However, since feature selection methods can be classified into filter, wrapper, and embedded methods [
23], none of them consider all three types of methods for missing value imputation, especially for medical datasets. Additionally, the numbers of features in their chosen datasets are also very small (i.e., 13 to 105 and 6 to 9).
Therefore, the research objective in this study is to examine the effects of performing three types of feature selection methods on model-based missing value imputation over different medical domain datasets. For feature selection, three different types of feature selection methods are employed; information gain (IG) as the filter-based method, genetic algorithm (GA) as the wrapper-based method, and decision tree (DT) as the embedded-based method. In addition, three popular machine learning techniques are used for the imputation process, namely the k-nearest neighbor (k-NN), multilayer perceptron (MLP), and support vector machine (SVM) approaches.
The contribution of this paper is two-fold. First, the effect of performing feature selection on missing value imputation is examined for various domain problems. Second, the best combinations of feature selection and imputation methods are identified for datasets with different dimensionality scales.
The rest of this paper is organized as follows. A review of the related literature is given in
Section 2, including the types of missing values and the missing value imputation process.
Section 3 describes the experimental procedure,
Section 4 presents the experimental results, and some conclusions are provided in
Section 5.
3. Research Methodology
3.1. Combination of Feature Selection and Missing Value Imputation
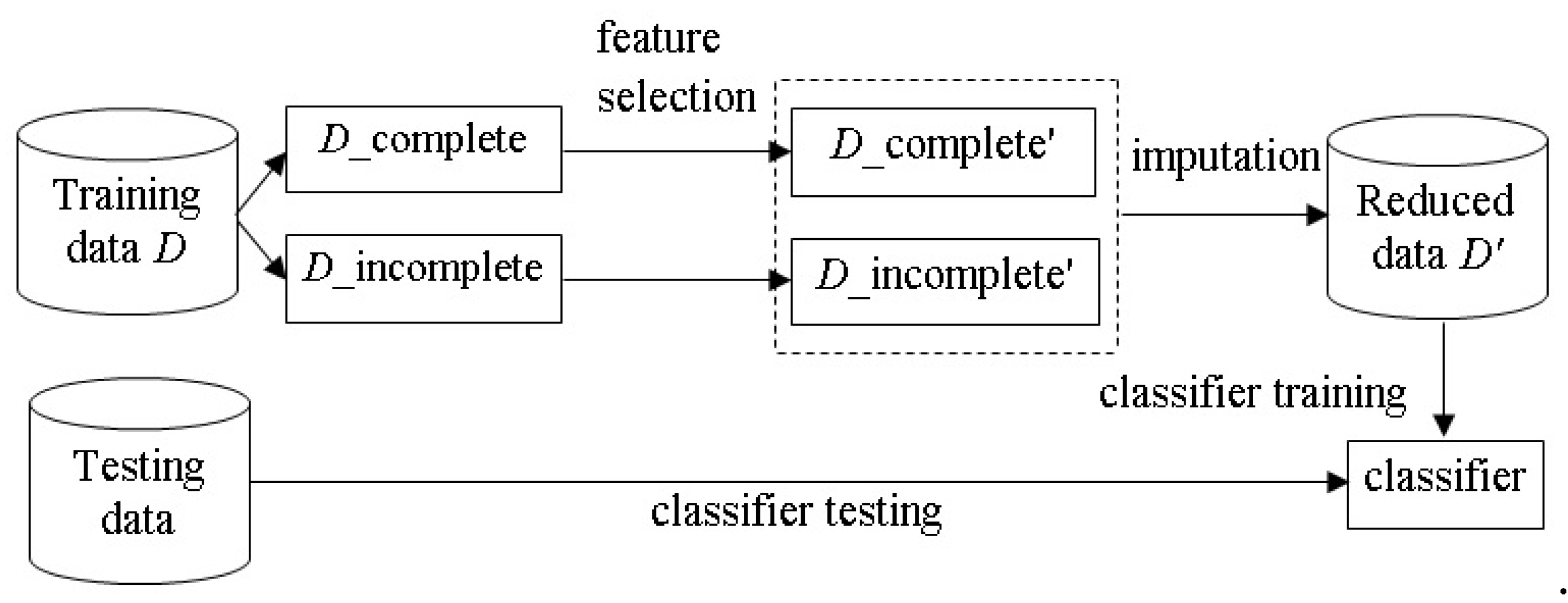
The process combining feature selection and missing value imputation is illustrated in
Figure 1. The incomplete
M dimension dataset
D is composed of training and test sets, denoted by
D_tr and
D_te, respectively. For feature selection,
D_tr contains a number of complete (i.e.,
D_complete) and incomplete (i.e.,
D_incomplete) data samples. The feature selection step is performed on the
D_complete subset, leading to a new subset that contains
N dimensions (where
N <
M), denoted as
D_complete’. It should be noted that the feature selection process only considers the data in
D_complete, since each of these data contains no missing attribute values, which allow feature selection algorithms to successfully select a subset of representative features. However, the issue of whether
D_complete represents the population is beyond the scope of this paper. Next, the
D_incomplete subset is also reduced to the same
N dimensional subset, denoted as
D_incomplete’.
For missing value imputation, the D_complete’ subset is used to construct a learning model. For the example of imputing the missing value of the i-th feature (i = 1, 2, …, N) in D_incomplete’, the learning model is trained by the data samples of D_incomplete’, where the i-th feature of D_incomplete’ is used as the output feature and the rest of the features are the input features. Then, estimations are produced by the model to replace the missing values in the D_incomplete’ subset.
The same process is performed over the testing set D_te. That is, the M dimensional testing set D_te is reduced to the N dimensional testing set, denoted by D_te’. Next, missing value imputation is performed by the learned model trained by D_complete’. Finally, the imputed dataset, denoted as reduced data D_tr’, is used to train a classifier, and its classification performance is examined by the reduced testing set (i.e., D_te’).
The baseline imputation process, without feature selection being performed, uses D_complete directly with the model to produce estimations for the missing values of D_incomplete. The aim of this study is to examine differences in performance between the combined feature selection and imputation method and the baseline imputation method.
3.2. Experimental Setup
The experiment is based on five UCI (University of California, Irvine) datasets, in which three datasets contain relatively lower dimensional features and the other two are of higher dimensions. Choosing these datasets with different feature dimensions leads to make the final conclusion. The basic information for the five datasets is listed in
Table 1.
For each dataset, missing values are simulated by the MCAR mechanism. The results of calculations with both imputation processes obtained with different missing rates, ranging from 10% to 50% at 10% intervals, are compared in order to understand the performance trends. Note that for larger missing rates with MCAR, each data sample in the training set is likely to become incomplete, which means that there is no data sample in the D_complete subset. Therefore, the criterion for performing the missing rate simulation is that at least 5 training data samples should be complete, without any missing values.
Moreover, each dataset is divided into 90% training and 10% testing datasets by the 10-fold cross validation method [
44]. The final classification performance of a classifier is based on the average of 10 test results. Specifically, for each missing rate, each of the 10-fold training sets is simulated 10 times, resulting in 100 different training sets under a specific missing rate. Finally, the feature selection and final classification performance is averaged by the 100 results in order to avoid the bias result produced by the MCAR mechanism.
Three feature selection algorithms are compared, namely information gain (IG), a type of filter method; the genetic algorithm (GA) as a type of wrapper method; and C4.5 decision tree (DT) as a type of embedded method. They are implemented using Weka software (
http://www.cs.waikato.ac.nz/ml/weka/). In particular, for the IG the feature selection method, the top ranked 50%, 65%, and 80% of features are kept and compared. Our results show that using the top ranked 80% of original features outperforms the other two settings. Therefore, we only report the best result of IG in this paper. For GA, the predictor and searcher functions are based on “WrapperSubsetEval” and “Genetic Search” functions in Weka software, respectively. For DT, the J48 decision tree classifier is used, where the nodes in the constructed tree are regarded as the selected features.
For missing value imputation, three deferent learning models are constructed, namely the k-nearest neighbor (KNN), multilayer perceptron (MLP) neural network, and support vector machine (SVM) models. As a result, there are nine different combinations of the three feature selection methods and three imputation models. Note that the parameters for constructing these models are based on the default parameters in Weka software. Note that since the aim of this paper is to examine whether performing feature selection can affect the imputation result and classification performance, tuning the parameters to find out the best classifier is not the research objective of this paper.
Finally, SVM is considered for classifier design, since it is the most widely used technique for pattern classification and has shown its effectiveness in many pattern recognition problems [
45].
5. Conclusions
Missing value imputation is a solution for the incomplete dataset problem. Given that the imputation process requires a set of observed data for imputation modeling, regardless of whether statistical or machine learning techniques are used to produce estimations to replace the missing values, the quality of the observed data is critical. In this paper, we focus on the problem from the feature selection perspective, assuming that some of the collected features may be unrepresentative and affect the imputation results, leading in turn to degradation of the final performance of the classifiers when compared with the ones where feature selection is performed.
For the experiments, five different medical domain datasets containing various numbers of feature dimensions are used. In addition, three different types of feature selection methods are compared, namely information gain (IG) as the filter method, genetic algorithm (GA) as the wrapper method, and decision tree (DT) as the embedded method. For missing value imputation, the multilayer perceptron (MLP) neural network, k-nearest neighbor (KNN), and support vector machine (SVM) models are constructed individually.
The experimental results show that the combination of feature selection and imputation can make the classifier (i.e., SVM) perform better than the baseline classifier without feature selection for many datasets with different missing rates. For lower dimensional datasets, using GA and IG for feature selection is recommended, whereas DT is a better choice for higher dimensional datasets.
Some issues should be considered in future research work. First, other missingness mechanisms, including MAR and MNAR, can be investigated for the feature selection effect. In addition, some datasets that naturally have specific numbers of missing data (i.e., specific missing rates) can be used. On the other hand, some other differences among the datasets that could influence the results can also be used, for example binary or multiple differences, or even the difficulty in classification where the datasets contain much higher dimensions or larger numbers of instances and classes. Second, in performing feature selection and missing value imputation, the major limitation is that a number of observed data (i.e.,
D_complete) must be provided for the feature selection methods to select some representative features and imputation models to produce estimations to replace the missing values. Therefore, the effect of using different numbers of observed data on the feature selection and imputation results should be investigated. On the other hand, for datasets that do not contain a sufficient number of complete data samples, the over-sampling techniques [
46,
47] used to create synthetic samples can be employed. Lastly, very high dimensional datasets in specific domain problems containing several hundreds of thousands of dimensions, such as text and sensor array data, should be further investigated to assess the level of impact of performing feature selection over very high dimensional incomplete datasets.