Glycan-Dependent and -Independent Dual Recognition between DC-SIGN and Type II Serine Protease MSPL/TMPRSS13 in Colorectal Cancer Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Preparation of Recombinant Proteins
2.2. DC-SIGN-Fc Affinity Chromatography
2.3. Co-Precipitation Assay, MSPL Protease Digestion Assay, and DC-SIGN Lectin Blot
2.4. Immunohistochemical Staining of COLO205 Cells and Human Colorectal Cancer Tissues
2.5. Flow Cytometry
3. Results
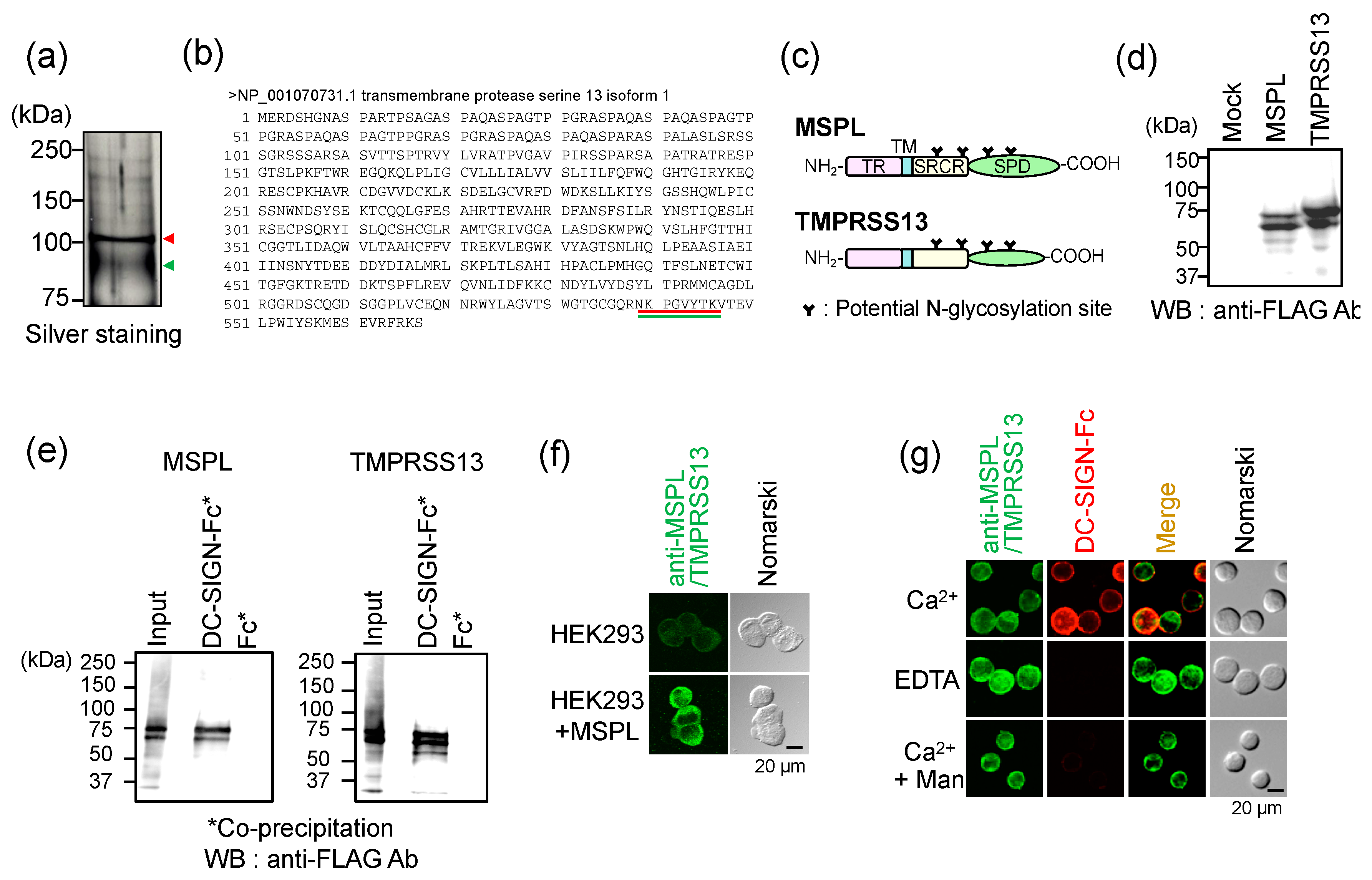
3.1. Identification of MSPL/TMPRSS13 as DC-SIGN Ligands
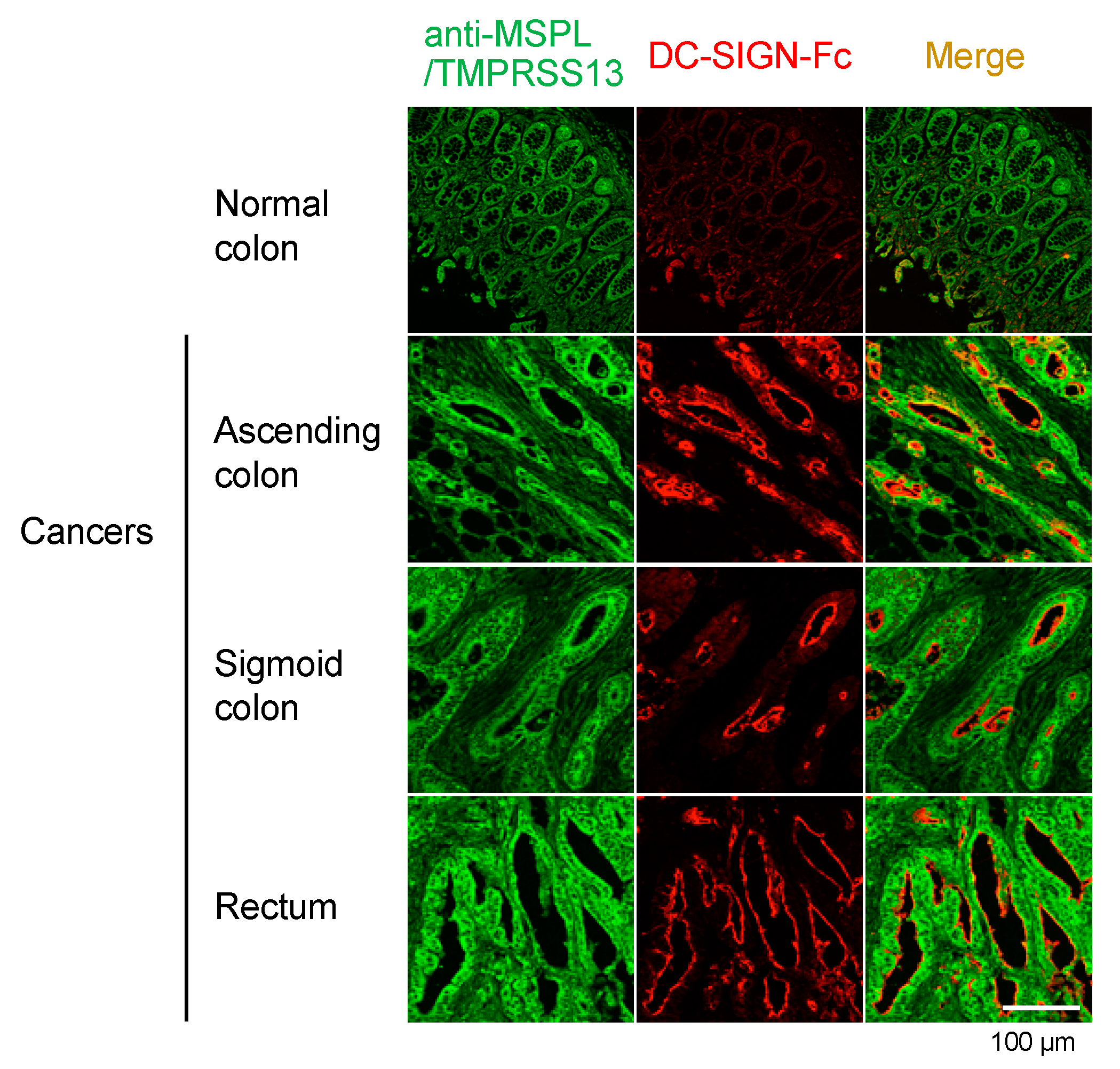
3.2. Localization of MSPL/TMPRSS13 in Colorectal Carcinoma Tissue
3.3. Glycan-Dependent Recognition of MSPL by DC-SIGN
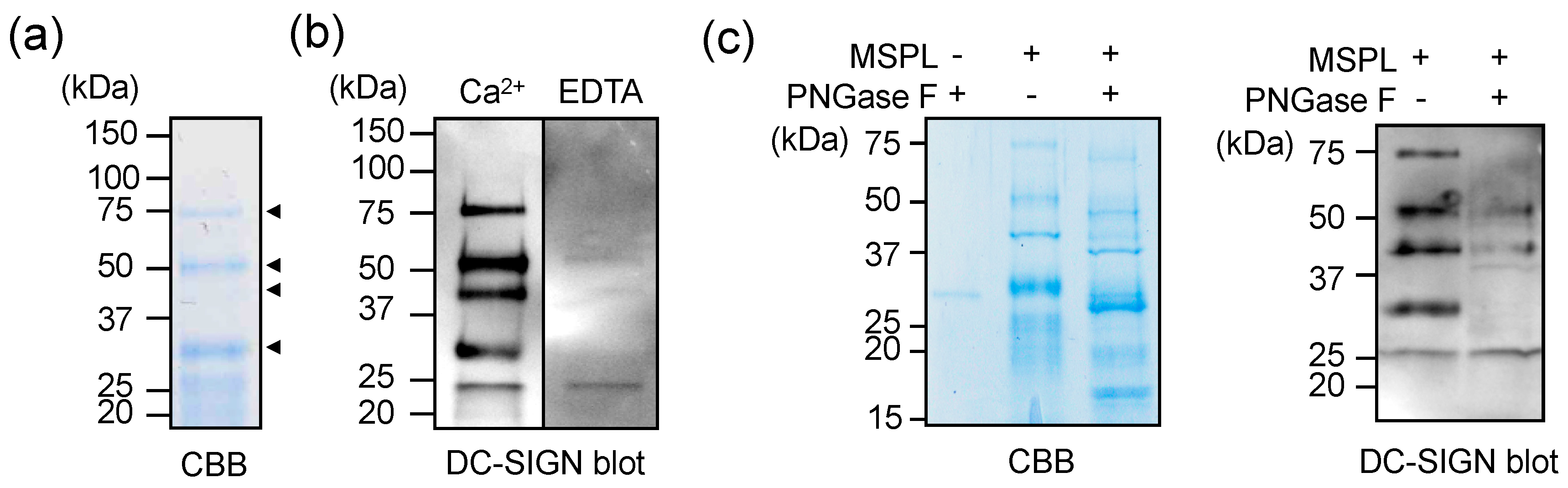
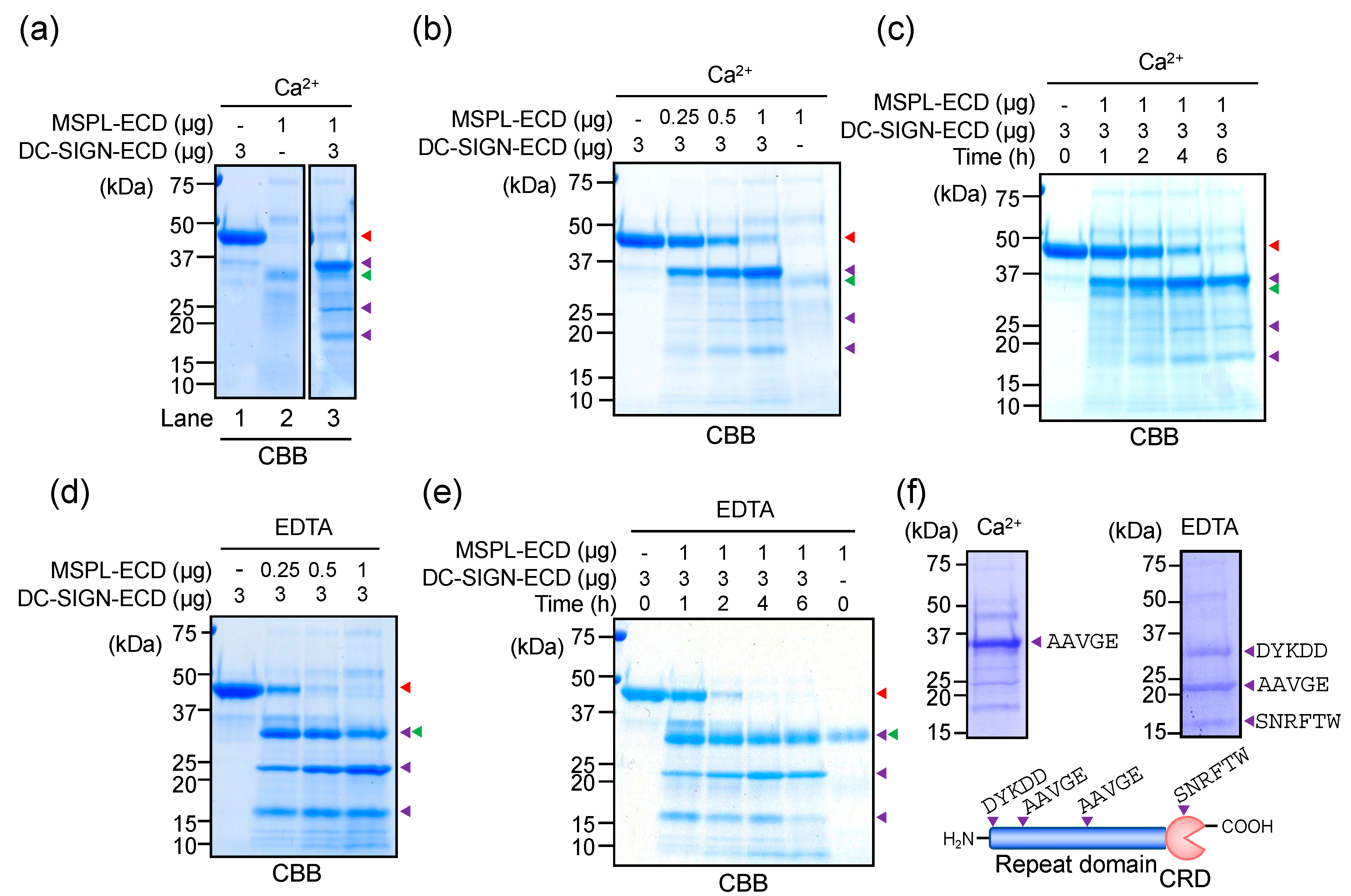
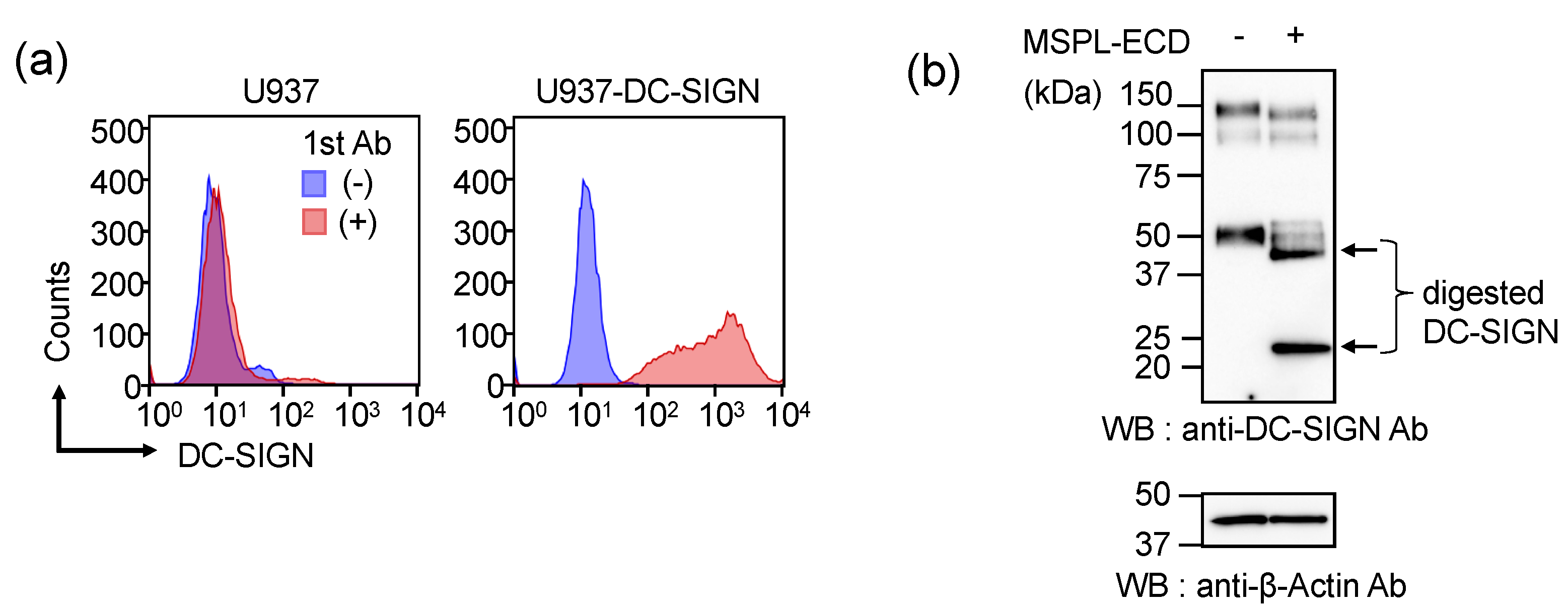
3.4. Glycan-Independent Digestion of DC-SIGN by MSPL
3.5. Cellular DC-SIGN as a Target of MSPL
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Geijtenbeek, T.B.; Torensma, R.; van Vliet, S.J.; van Duijnhoven, G.C.; Adema, G.J.; van Kooyk, Y.; Figdor, C.G. Identification of DC-SIGN, a novel dendritic cell-specific ICAM-3 receptor that supports primary immune responses. Cell 2000, 100, 575–585. [Google Scholar] [CrossRef]
- Feinberg, H.; Mitchell, D.A.; Drickamer, K.; Weis, W.I. Structural basis for selective recognition of oligosaccharides by DC-SIGN and DC-SIGNR. Science 2001, 294, 2163–2166. [Google Scholar] [CrossRef] [PubMed]
- Jameson, B.; Baribaud, F.; Pohlmann, S.; Ghavimi, D.; Mortari, F.; Doms, R.W.; Iwasaki, A. Expression of DC-SIGN by Dendritic Cells of Intestinal and Genital Mucosae in Humans and Rhesus Macaques. J. Virol. 2002, 76, 1866–1875. [Google Scholar] [CrossRef] [PubMed]
- Fehres, C.M.; van Beelen, A.J.; Bruijns, S.C.M.; Ambrosini, M.; Kalay, H.; van Bloois, L.; Unger, W.W.J.; Garcia-Vallejo, J.J.; Storm, G.; de Gruijl, T.D.; et al. In situ Delivery of Antigen to DC-SIGN + CD14 + Dermal Dendritic Cells Results in Enhanced CD8 + T-Cell Responses. J. Investig. Dermatol. 2015, 135, 2228–2236. [Google Scholar] [CrossRef] [PubMed]
- Geijtenbeek, T.B.; Kwon, D.S.; Torensma, R.; van Vliet, S.J.; van Duijnhoven, G.C.; Middel, J.; Cornelissen, I.L.; Nottet, H.S.; KewalRamani, V.N.; Littman, D.R.; et al. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell 2000, 100, 587–597. [Google Scholar] [CrossRef]
- Lai, W.K.; Sun, P.J.; Zhang, J.; Jennings, A.; Lalor, P.F.; Hubscher, S.; McKeating, J.A.; Adams, D.H. Expression of DC-SIGN and DC-SIGNR on human sinusoidal endothelium: A role for capturing hepatitis C virus particles. Am. J. Pathol. 2006, 169, 200–208. [Google Scholar] [CrossRef]
- Sakuntabhai, A.; Turbpaiboon, C.; Casadémont, I.; Chuansumrit, A.; Lowhnoo, T.; Kajaste-Rudnitski, A.; Kalayanarooj, S.M.; Tangnararatchakit, K.; Tangthawornchaikul, N.; Vasanawathana, S.; et al. A variant in the CD209 promoter is associated with severity of dengue disease. Nat. Genet. 2005, 37, 507–513. [Google Scholar] [CrossRef]
- Geijtenbeek, T.B.H.; Van Vliet, S.J.; Koppel, E.A.; Sanchez-Hernandez, M.; Vandenbroucke-Grauls, C.M.J.E.; Appelmelk, B.; Van Kooyk, Y. Mycobacteria target DC-SIGN to suppress dendritic cell function. J. Exp. Med. 2003, 197, 7–17. [Google Scholar] [CrossRef]
- Tailleux, L.; Schwartz, O.; Herrmann, J.-L.; Pivert, E.; Jackson, M.; Amara, A.; Legres, L.; Dreher, D.; Nicod, L.P.; Gluckman, J.C.; et al. DC-SIGN is the major Mycobacterium tuberculosis receptor on human dendritic cells. J. Exp. Med. 2003, 197, 121–127. [Google Scholar] [CrossRef]
- Maeda, N.; Nigou, J.; Herrmann, J.-L.; Jackson, M.; Amara, A.; Lagrange, P.H.; Puzo, G.; Gicquel, B.; Neyrolles, O. The Cell Surface Receptor DC-SIGN Discriminates between Mycobacterium Species through Selective Recognition of the Mannose Caps on Lipoarabinomannan. J. Biol. Chem. 2003, 278, 5513–5516. [Google Scholar] [CrossRef]
- Colmenares, M.; Puig-Kröger, A.; Pello, O.M.; Corbí, A.L.; Rivas, L. Dendritic cell (DC)-specific intercellular adhesion molecule 3 (ICAM-3)-grabbing nonintegrin (DC-SIGN, CD209), a C-type surface lectin in human DCs, is a receptor for Leishmania amastigotes. J. Biol. Chem. 2002, 277, 36766–36769. [Google Scholar] [CrossRef] [PubMed]
- Kwon, D.S.; Gregorio, G.; Bitton, N.; Hendrickson, W.A.; Littman, D.R. DC-SIGN-Mediated Internalization of HIV Is Required for Trans-Enhancement of T Cell Infection. Immunity 2002, 16, 135–144. [Google Scholar] [CrossRef]
- Vannberg, F.O.; Chapman, S.J.; Khor, C.C.; Tosh, K.; Floyd, S.; Jackson-Sillah, D.; Crampin, A.; Sichali, L.; Bah, B.; Gustafson, P.; et al. CD209 Genetic Polymorphism and Tuberculosis Disease. PLoS ONE 2008, 3, e1388. [Google Scholar] [CrossRef] [PubMed]
- Den Dunnen, J.; Gringhuis, S.I.; Geijtenbeek, T.B.H. Innate signaling by the C-type lectin DC-SIGN dictates immune responses. Cancer Immunol. Immunother. 2009, 58, 1149–1157. [Google Scholar] [CrossRef] [PubMed]
- Gringhuis, S.I.; den Dunnen, J.; Litjens, M.; van het Hof, B.; van Kooyk, Y.; Geijtenbeek, T.B.H. C-Type Lectin DC-SIGN Modulates Toll-like Receptor Signaling via Raf-1 Kinase-Dependent Acetylation of Transcription Factor NF-κB. Immunity 2007, 26, 605–616. [Google Scholar] [CrossRef] [PubMed]
- Van Gisbergen, K.P.J.M.; Aarnoudse, C.A.; Meijer, G.A.; Geijtenbeek, T.B.H.; van Kooyk, Y. Dendritic cells recognize tumor-specific glycosylation of carcinoembryonic antigen on colorectal cancer cells through dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin. Cancer Res. 2005, 65, 5935–5944. [Google Scholar] [CrossRef]
- Nonaka, M.; Ma, B.Y.; Murai, R.; Nakamura, N.; Baba, M.; Kawasaki, N.; Hodohara, K.; Asano, S.; Kawasaki, T. Glycosylation-dependent interactions of C-type lectin DC-SIGN with colorectal tumor-associated Lewis glycans impair the function and differentiation of monocyte-derived dendritic cells. J. Immunol. 2008, 180, 3347–3356. [Google Scholar] [CrossRef]
- Nonaka, M.; Ma, B.Y.; Imaeda, H.; Kawabe, K.; Kawasaki, N.; Hodohara, K.; Kawasaki, N.; Andoh, A.; Fujiyama, Y.; Kawasaki, T. Dendritic cell-specific intercellular adhesion molecule 3-grabbing non-integrin (DC-SIGN) recognizes a novel ligand, Mac-2-binding protein, characteristically expressed on human colorectal carcinomas. J. Biol. Chem. 2011, 286, 22403–22413. [Google Scholar] [CrossRef]
- Kim, D.R.; Sharmin, S.; Inoue, M.; Kido, H. Cloning and expression of novel mosaic serine proteases with and without a transmembrane domain from human lung. Biochim. Biophys. Acta 2001, 1518, 204–209. [Google Scholar] [CrossRef]
- Bugge, T.H.; Antalis, T.M.; Wu, Q. Type II transmembrane serine proteases. J. Biol. Chem. 2009, 284, 23177–23181. [Google Scholar] [CrossRef]
- Szabo, R.; Bugge, T.H. Membrane-anchored serine proteases in vertebrate cell and developmental biology. Annu. Rev. Cell Dev. Biol. 2011, 27, 213–235. [Google Scholar] [CrossRef] [PubMed]
- Antalis, T.M.; Buzza, M.S.; Hodge, K.M.; Hooper, J.D.; Netzel-Arnett, S. The cutting edge: Membrane-anchored serine protease activities in the pericellular microenvironment. Biochem. J. 2010, 428, 325–346. [Google Scholar] [CrossRef] [PubMed]
- Madsen, D.H.; Szabo, R.; Molinolo, A.A.; Bugge, T.H. TMPRSS13 deficiency impairs stratum corneum formation and epidermal barrier acquisition. Biochem. J. 2014, 461, 487–495. [Google Scholar] [CrossRef] [PubMed]
- Murray, A.S.; Varela, F.A.; Hyland, T.E.; Schoenbeck, A.J.; White, J.M.; Tanabe, L.M.; Todi, S.V.; List, K. Phosphorylation of the type II transmembrane serine protease, TMPRSS13, in hepatocyte growth factor activator inhibitor-1 and -2-mediated cell-surface localization. J. Biol. Chem. 2017, 292, 14867–14884. [Google Scholar] [CrossRef]
- Kido, H.; Okumura, Y. MSPL/TMPRSS13. Front. Biosci. 2008, 13, 754–758. [Google Scholar] [CrossRef]
- Tanabe, L.M.; List, K. The role of type II transmembrane serine protease-mediated signaling in cancer. FEBS J. 2017, 284, 1421–1436. [Google Scholar] [CrossRef]
- Louvard, D.; Maroux, S.; Baratti, J.; Desnuelle, P. On the distribution of enterokinase in porcine intestine and on its subcellular localization. Biochim. Biophys. Acta 1973, 309, 127–137. [Google Scholar] [CrossRef]
- Lin, C.Y.; Anders, J.; Johnson, M.; Dickson, R.B. Purification and characterization of a complex containing matriptase and a Kunitz-type serine protease inhibitor from human milk. J. Biol. Chem. 1999, 274, 18237–18242. [Google Scholar] [CrossRef]
- Yamaoka, K.; Masuda, K.; Ogawa, H.; Takagi, K.; Umemoto, N.; Yasuoka, S. Cloning and characterization of the cDNA for human airway trypsin-like protease. J. Biol. Chem. 1998, 273, 11895–11901. [Google Scholar] [CrossRef]
- Afar, D.E.; Vivanco, I.; Hubert, R.S.; Kuo, J.; Chen, E.; Saffran, D.C.; Raitano, A.B.; Jakobovits, A. Catalytic cleavage of the androgen-regulated TMPRSS2 protease results in its secretion by prostate and prostate cancer epithelia. Cancer Res. 2001, 61, 1686–1692. [Google Scholar]
- Zmora, P.; Blazejewska, P.; Moldenhauer, A.-S.; Welsch, K.; Nehlmeier, I.; Wu, Q.; Schneider, H.; Pöhlmann, S.; Bertram, S. DESC1 and MSPL activate influenza A viruses and emerging coronaviruses for host cell entry. J. Virol. 2014, 88, 12087–12097. [Google Scholar] [CrossRef]
- Kido, H.; Okumura, Y.; Takahashi, E.; Pan, H.-Y.; Wang, S.; Chida, J.; Le, T.Q.; Yano, M. Host envelope glycoprotein processing proteases are indispensable for entry into human cells by seasonal and highly pathogenic avian influenza viruses. J. Mol. Genet Med. 2008, 3, 167–175. [Google Scholar] [CrossRef]
- Okumura, Y.; Takahashi, E.; Yano, M.; Ohuchi, M.; Daidoji, T.; Nakaya, T.; Böttcher, E.; Garten, W.; Klenk, H.-D.; Kido, H. Novel type II transmembrane serine proteases, MSPL and TMPRSS13, Proteolytically activate membrane fusion activity of the hemagglutinin of highly pathogenic avian influenza viruses and induce their multicycle replication. J. Virol. 2010, 84, 5089–5096. [Google Scholar] [CrossRef]
- Webb, S.L.; Sanders, A.J.; Mason, M.D.; Jiang, W.G. Type II transmembrane serine protease (TTSP) deregulation in cancer. Front Biosci 2011, 16, 539–552. [Google Scholar] [CrossRef]
- Wu, Q.; Parry, G. Hepsin and prostate cancer. Front. Biosci. 2007, 12, 5052–5059. [Google Scholar] [CrossRef]
- Tanimoto, H.; Yan, Y.; Clarke, J.; Korourian, S.; Shigemasa, K.; Parmley, T.H.; Parham, G.P.; O’Brien, T.J. Hepsin, a cell surface serine protease identified in hepatoma cells, is overexpressed in ovarian cancer. Cancer Res. 1997, 57, 2884–2887. [Google Scholar]
- Holt, S.K.; Kwon, E.M.; Lin, D.W.; Ostrander, E.A.; Stanford, J.L. Association of hepsin gene variants with prostate cancer risk and prognosis. Prostate 2010, 70, 1012–1019. [Google Scholar] [CrossRef]
- Matsuo, T.; Nakamura, K.; Takamoto, N.; Kodama, J.; Hongo, A.; Abrzua, F.; Nasu, Y.; Kumon, H.; Hiramatsu, Y. Expression of the serine protease hepsin and clinical outcome of human endometrial cancer. Anticancer Res. 2008, 28, 159–164. [Google Scholar]
- Hakomori, S. Antigen structure and genetic basis of histo-blood groups A, B and O: Their changes associated with human cancer. Biochim. Biophys. Acta 1999, 1473, 247–266. [Google Scholar] [CrossRef]
- Terada, M.; Khoo, K.-H.; Inoue, R.; Chen, C.-I.; Yamada, K.; Sakaguchi, H.; Kadowaki, N.; Ma, B.Y.; Oka, S.; Kawasaki, T.; et al. Characterization of oligosaccharide ligands expressed on SW1116 cells recognized by mannan-binding protein. A highly fucosylated polylactosamine type N-glycan. J. Biol. Chem. 2005, 280, 10897–10913. [Google Scholar] [CrossRef]
- Kawasaki, N.; Lin, C.-W.; Inoue, R.; Khoo, K.-H.; Kawasaki, N.; Ma, B.Y.; Oka, S.; Ishiguro, M.; Sawada, T.; Ishida, H.; et al. Highly fucosylated N-glycan ligands for mannan-binding protein expressed specifically on CD26 (DPPIV) isolated from a human colorectal carcinoma cell line, SW1116. Glycobiology 2009, 19, 437–450. [Google Scholar] [CrossRef] [PubMed]
- Nonaka, M.; Imaeda, H.; Matsumoto, S.; Yong Ma, B.; Kawasaki, N.; Mekata, E.; Andoh, A.; Saito, Y.; Tani, T.; Fujiyama, Y.; et al. Mannan-binding protein, a C-type serum lectin, recognizes primary colorectal carcinomas through tumor-associated Lewis glycans. J. Immunol. 2014, 192, 1294–1301. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nonaka, M.; Matsumoto, S.; Ma, B.Y.; Kido, H.; Kawasaki, N.; Kawasaki, N.; Kawasaki, T. Glycan-Dependent and -Independent Dual Recognition between DC-SIGN and Type II Serine Protease MSPL/TMPRSS13 in Colorectal Cancer Cells. Appl. Sci. 2020, 10, 2687. https://doi.org/10.3390/app10082687
Nonaka M, Matsumoto S, Ma BY, Kido H, Kawasaki N, Kawasaki N, Kawasaki T. Glycan-Dependent and -Independent Dual Recognition between DC-SIGN and Type II Serine Protease MSPL/TMPRSS13 in Colorectal Cancer Cells. Applied Sciences. 2020; 10(8):2687. https://doi.org/10.3390/app10082687
Chicago/Turabian StyleNonaka, Motohiro, Shogo Matsumoto, Bruce Yong Ma, Hiroshi Kido, Nana Kawasaki, Nobuko Kawasaki, and Toshisuke Kawasaki. 2020. "Glycan-Dependent and -Independent Dual Recognition between DC-SIGN and Type II Serine Protease MSPL/TMPRSS13 in Colorectal Cancer Cells" Applied Sciences 10, no. 8: 2687. https://doi.org/10.3390/app10082687
APA StyleNonaka, M., Matsumoto, S., Ma, B. Y., Kido, H., Kawasaki, N., Kawasaki, N., & Kawasaki, T. (2020). Glycan-Dependent and -Independent Dual Recognition between DC-SIGN and Type II Serine Protease MSPL/TMPRSS13 in Colorectal Cancer Cells. Applied Sciences, 10(8), 2687. https://doi.org/10.3390/app10082687



