Preliminary Study to Explore the Immune-Enhancement Mechanism of Platycodon grandiflorus Extract through Comparative Transcriptome Analysis
Abstract
:Featured Application
Abstract
1. Introduction
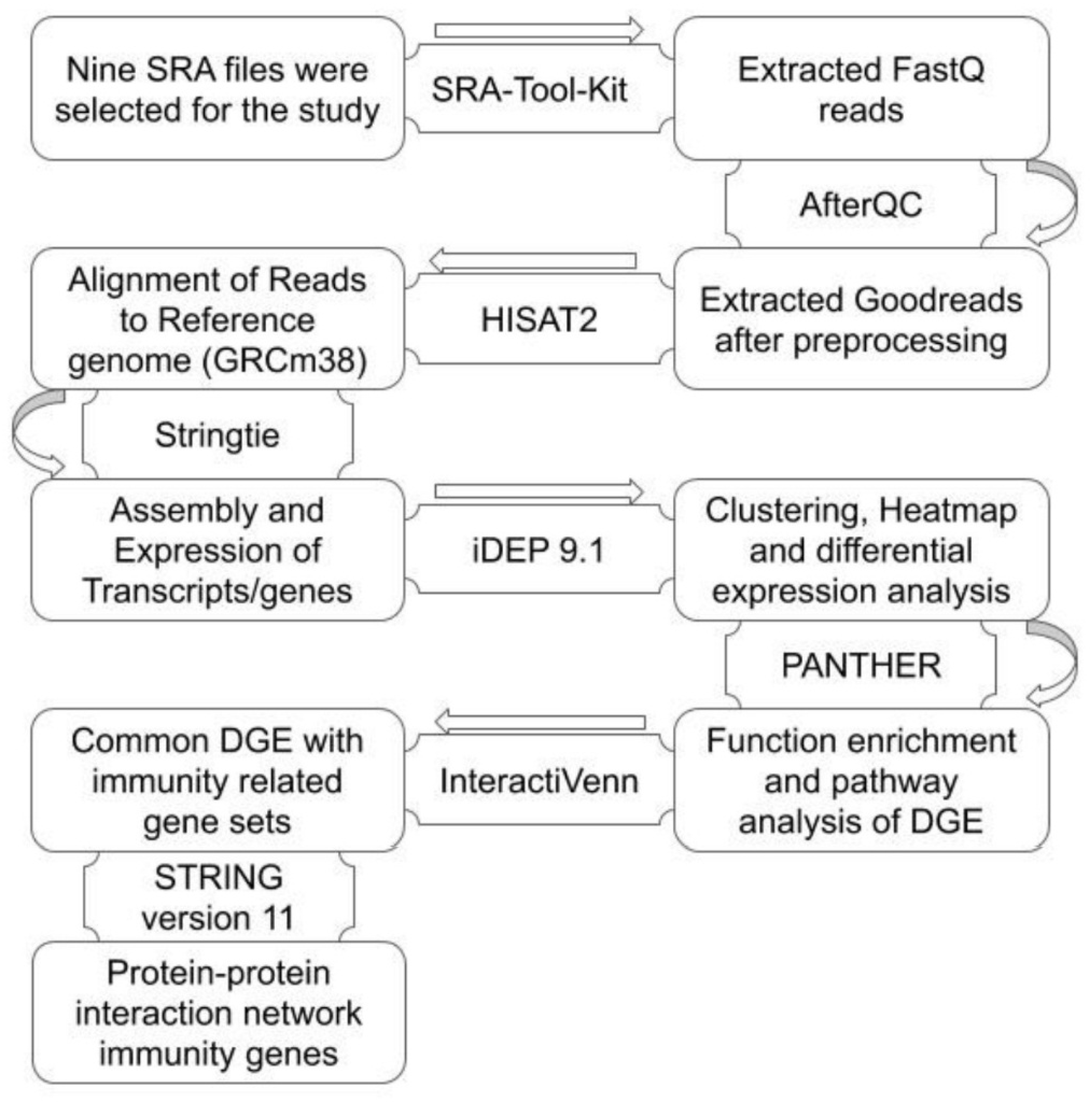
2. Materials and Methods
2.1. RNA-Seq Data Sets Used in the Study
2.2. RNA-Seq Analysis for Assembly and Differential Expression
2.3. Function Enrichment of DEG
2.4. Identification of DEG with Immunity Related Function in Subset Analysis
2.5. Protein-Protein Interaction Network
3. Results
3.1. Preprocessing and Alignment of Reads to Reference Genome
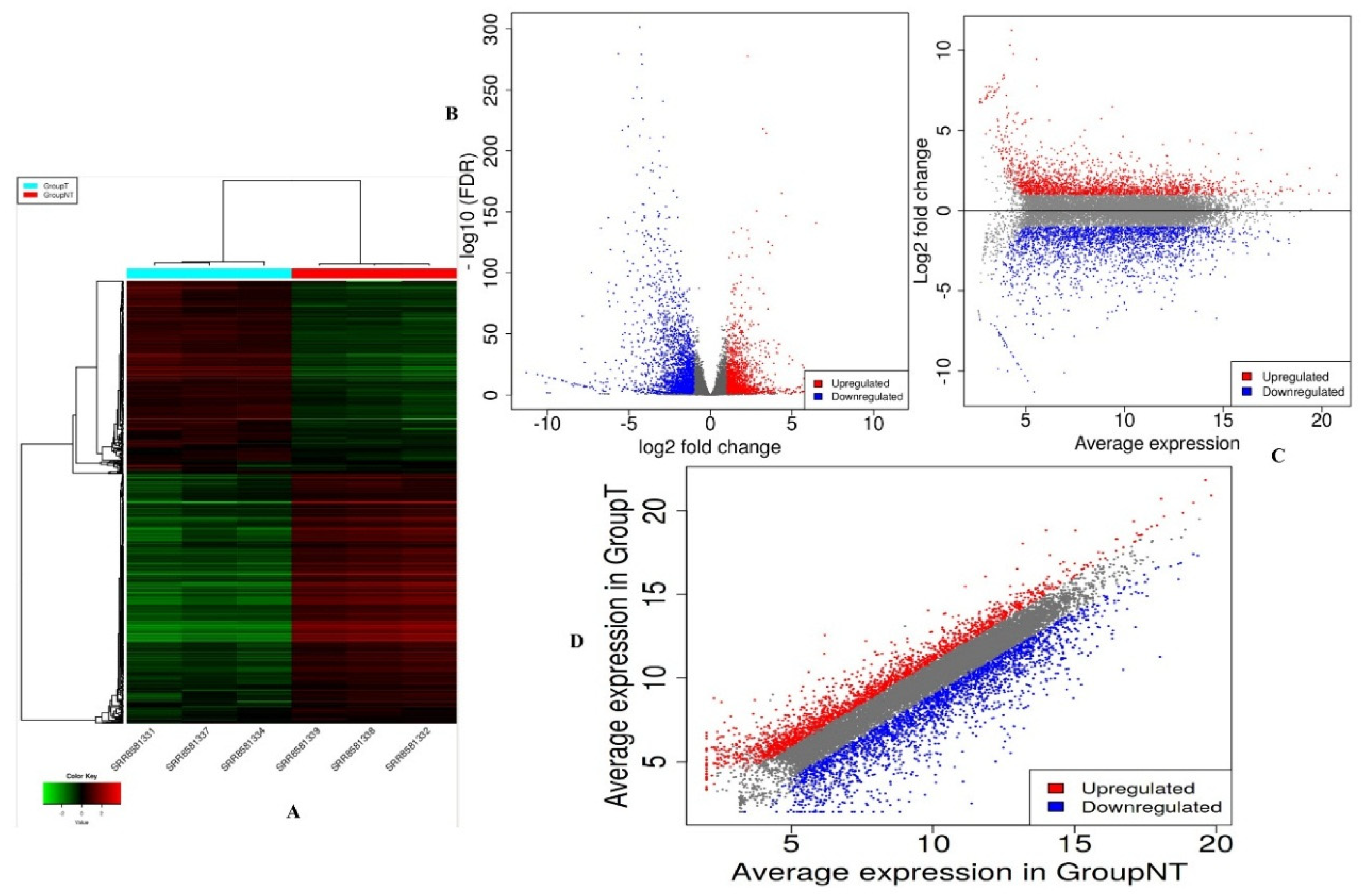
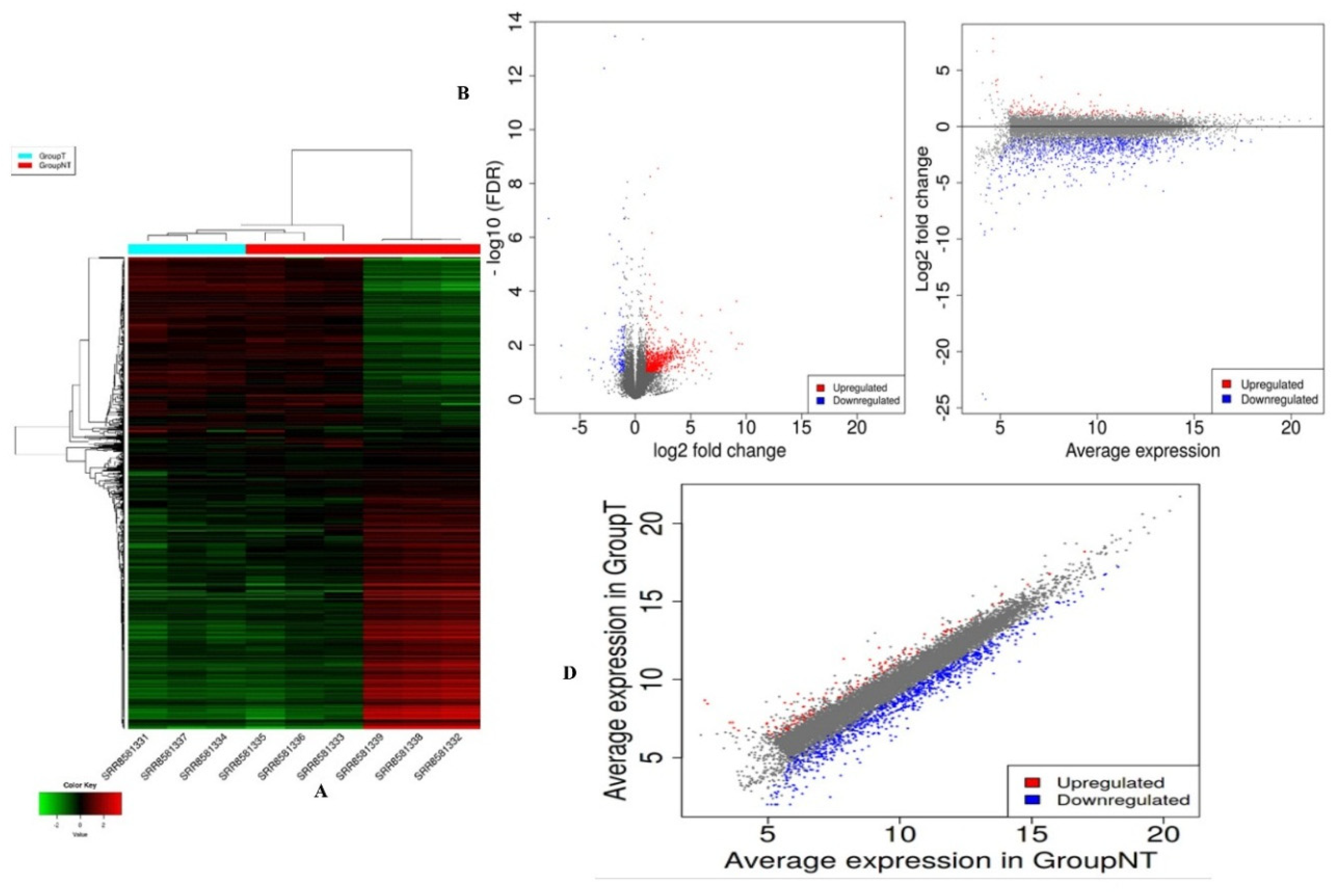
3.2. Assembly and Expression Analysis
3.3. Functional Enrichment Analysis
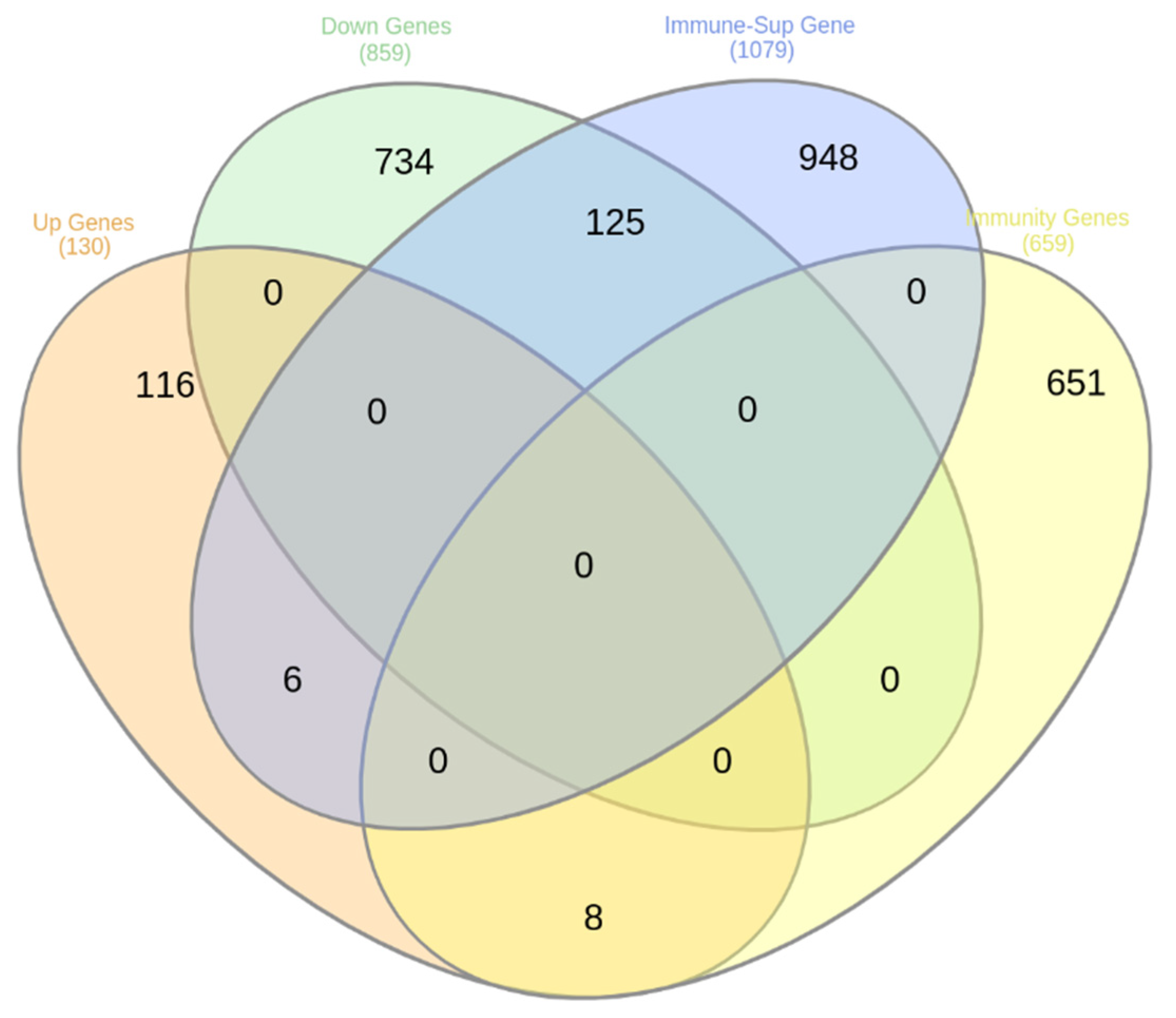
3.4. DEG with Immunity Related Function
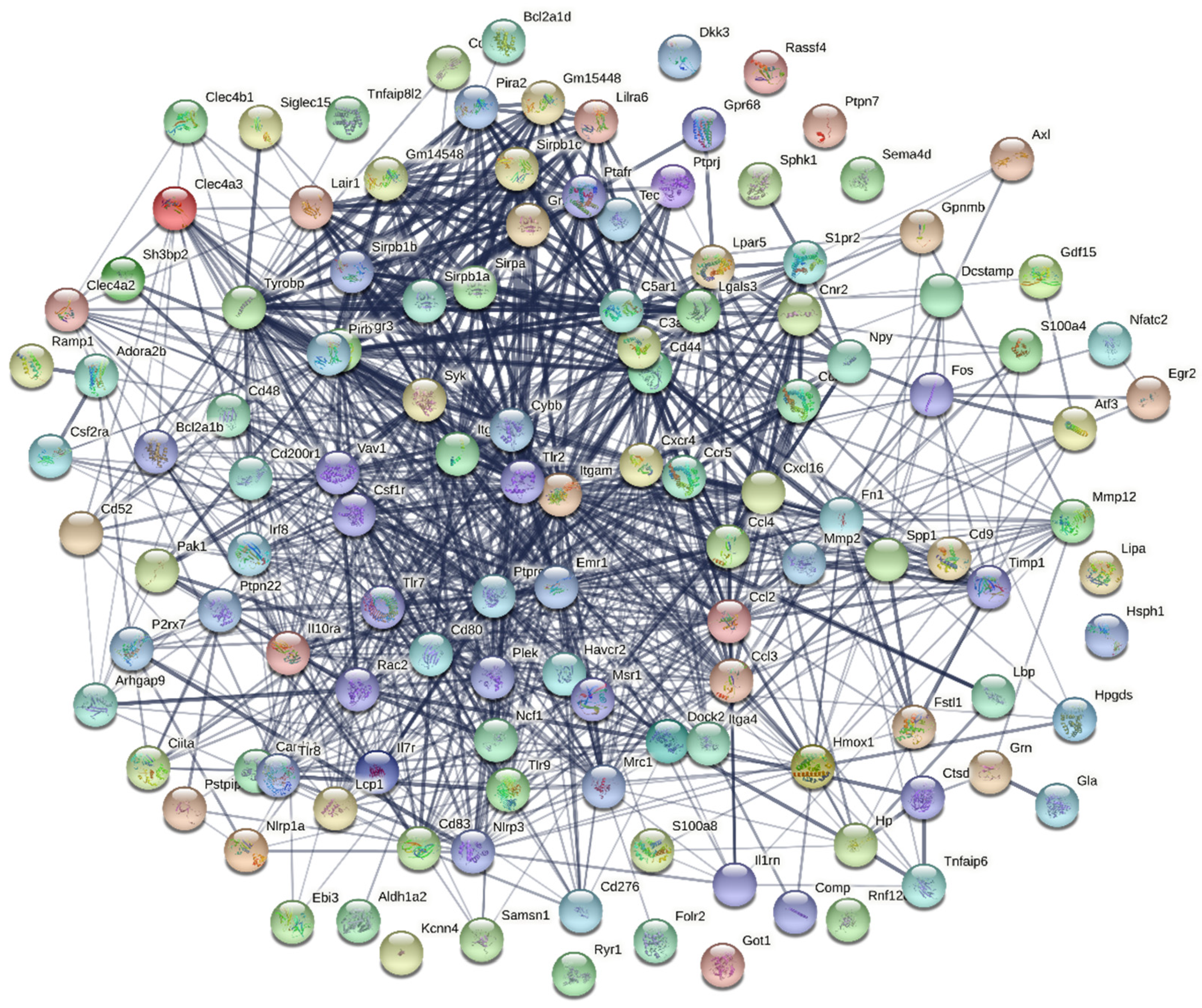
3.5. Protein–Protein Interaction Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ji, M.; Agula, B.; Yang, M.; Xu, J.; Jiang, L.-L.; Zhou, B.-C.; Li, M. The Pharmacological Effects and Health Benefits of Platycodon grandiflorus—A Medicine Food Homology Species. Foods 2020, 9, 142. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.-Y.; Hwang, W.-I.; Lim, S.-T. Antioxidant and anticancer activities of organic extracts from Platycodon grandiflorum A. De Candolle roots. J. Ethnopharmacol. 2004, 93, 409–415. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhao, M.; Zheng, W.; Liu, Y. Platycodin D, a triterpenoid saponin from Platycodon grandiflorum, suppresses the growth and invasion of human oral squamous cell carcinoma cells via the NF-κB pathway. J. Biochem. Mol. Toxicol. 2017, 31, e21934. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.J.; Kim, J.Y.; Choi, J.H.; Kim, H.G.; Chung, Y.C.; Roh, S.H.; Jeong, H.G. Inhibition of tumor invasion and metastasis by aqueous extract of the radix of Platycodon grandiflorum. Food Chem. Toxicol. 2006, 44, 1890–1896. [Google Scholar] [CrossRef] [PubMed]
- Park, E.-J.; Lee, H.-J. Immunomodulatory effects of fermented Platycodon grandiflorum extract through NF-κB signaling in RAW 264.7 cells. Nutrition Res. Prac. 2020, 14, 453–462. [Google Scholar] [CrossRef] [PubMed]
- Park, E.-J.; Lee, Y.-S.; Kim, S.M.; Jung, A.J.; Yoo, J.-H.; Lee, S.-H.; Jeong, H.C.; Lee, H.-J. Immune-Enhancing Effects of Red Platycodon grandiflorus Root Extract via p38 MAPK-Mediated NF-κB Activation. Appl. Sci. 2020, 10, 5457. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, Y.; Yan, P.; Cheng, G.; Wang, C.; Geng, N.; Wang, X.; Liu, J. Effects of Polysaccharides from Platycodon grandiflorum on Immunity-Enhancing Activity In Vitro. Molecules 2017, 22, 1918. [Google Scholar] [CrossRef] [Green Version]
- Zheng, P.; Fan, W.; Wang, S.; Hao, P.; Wang, Y.; Wan, H.; Hao, Z.; Liu, J.; Zhao, X. Characterization of polysaccharides extracted from Platycodon grandiflorus (Jacq.) A.DC. affecting activation of chicken peritoneal macrophages. Int. J. Biol. Macromol. 2017, 96, 775–785. [Google Scholar] [CrossRef]
- Noh, E.-M.; Kim, J.-M.; Lee, H.Y.; Song, H.-K.; Joung, S.O.; Yang, H.J.; Kim, M.J.; Kim, K.S.; Lee, Y.-R. Immuno-enhancement effects of Platycodon grandiflorum extracts in splenocytes and a cyclophosphamide-induced immunosuppressed rat model. BMC Complement. Altern. Med. 2019, 19, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Hamzić, E.; Kjaerup, R.B.; Mach, N.; Minozzi, G.; Strozzi, F.; Gualdi, V.; Williams, J.L.; Chen, J.; Wattrang, E.; Buitenhuis, B.; et al. RNA sequencing-based analysis of the spleen transcriptome following infectious bronchitis virus infection of chickens selected for different mannose-binding lectin serum concentrations. BMC Genom. 2016, 17, 1–13. [Google Scholar] [CrossRef]
- Sudhagar, A.; Kumar, G.; El-Matbouli, M. Transcriptome Analysis Based on RNA-Seq in Understanding Pathogenic Mechanisms of Diseases and the Immune System of Fish: A Comprehensive Review. Int. J. Mol. Sci. 2018, 19, 245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guleria, V.; Jaiswal, V. Comparative transcriptome analysis of different stages of Plasmodium falciparum to explore vaccine and drug candidates. Genomics 2020, 112, 796–804. [Google Scholar] [CrossRef] [PubMed]
- Lopez, B.I.; Santiago, K.G.; Lee, D.; Ha, S.; Seo, K. RNA Sequencing (RNA-Seq) Based Transcriptome Analysis in Immune Response of Holstein Cattle to Killed Vaccine against Bovine Viral Diarrhea Virus Type, I. Animals 2020, 10, 344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, P.; Jaiswal, V.; Pal, T.; Singh, J.; Chauhan, R.S. Comparative whole-transcriptome analysis in Podophyllum species identifies key transcription factors contributing to biosynthesis of podophyllotoxin in P. hexandrum. Protoplasma 2016, 254, 217–228. [Google Scholar] [CrossRef]
- Chawla, A.; Stobdan, T.; Srivastava, R.B.; Jaiswal, V.; Chauhan, R.S.; Kant, A. Sex-Biased Temporal Gene Expression in Male and Female Floral Buds of Seabuckthorn (Hippophae rhamnoides). PLoS ONE 2015, 10, e0124890. [Google Scholar] [CrossRef] [Green Version]
- Sood, A.; Jaiswal, V.; Chanumolu, S.K.; Malhotra, N.; Pal, T.; Chauhan, R.S. Mining whole genomes and transcriptomes of Jatropha (Jatropha curcas) and Castor bean (Ricinus communis) for NBS-LRR genes and defense response associated transcription factors. Mol. Biol. Rep. 2014, 41, 7683–7695. [Google Scholar] [CrossRef]
- Qi, Y.; Bin Liu, Y.; Rong, W.-H. RNA-Seq and its applications: A new technology for transcriptomics. Hereditas 2011, 33, 1191–1202. [Google Scholar] [CrossRef]
- Kim, Y.J.; Ryu, R.; Choi, J.-Y.; Choi, M.S. Platycodon grandiflorus Root Ethanol Extract Induces Lipid Excretion, Lipolysis, and Thermogenesis in Diet-Induced Obese Mice. J. Med. Food 2019, 22, 1100–1109. [Google Scholar] [CrossRef]
- Sherry, S.; Xiao, C.; Durbrow, K.; Kimelman, M.; Rodarmer, K.; Shumway, M.; Yaschenko, E. Ncbi sra toolkit technology for next generation sequence data. In Proceedings of the Plant and Animal Genome XX Conference, San Diego, CA, USA, 14–18 January 2012. [Google Scholar]
- Chen, S.; Huang, T.; Zhou, Y.; Han, Y.; Xu, M.; Gu, J. AfterQC: Automatic filtering, trimming, error removing and quality control for fastq data. BMC Bioinform. 2017, 18, 91–100. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Son, E.W.; Yao, R. iDEP: An integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinform. 2018, 19, 534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformation 2009, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Thomas, P.D.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019, 47, D419–D426. [Google Scholar] [CrossRef]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [Green Version]
- Giudicelli, V.; Chaume, D.; Lefranc, M.-P. IMGT/GENE-DB: A comprehensive database for human and mouse immunoglobulin and T cell receptor genes. Nucleic Acids Res. 2004, 33, D256–D261. [Google Scholar] [CrossRef]
- Liu, Y.; He, M.; Wang, D.; Diao, L.; Liu, J.; Tang, L.; Guo, S.; He, F.; Li, D. HisgAtlas 1.0: A human immunosuppression gene database. Database 2017, 2017. [Google Scholar] [CrossRef]
- Heberle, H.; Meirelles, G.V.; Da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 1–7. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Sun, J.; Liu, L.N.; Flies, D.B.; Nie, X.; Toki, M.; Zhang, J.; Song, C.; Zarr, M.; Zhou, X.; et al. Siglec-15 as an immune suppressor and potential target for normalization cancer immunotherapy. Nat. Med. 2019, 25, 656–666. [Google Scholar] [CrossRef] [PubMed]
- Kumar, H.; Kawai, T.; Akira, S. Toll-like receptors and innate immunity. Biochem. Biophys. Res. Commun. 2009, 388, 621–625. [Google Scholar] [CrossRef] [PubMed]
- Bhat, M.Y.; Solanki, H.S.; Advani, J.; Khan, A.A.; Prasad, T.S.K.; Gowda, H.; Thiyagarajan, S.; Chatterjee, A. Comprehensive network map of interferon gamma signaling. J. Cell Commun. Signal. 2018, 12, 745–751. [Google Scholar] [CrossRef] [PubMed]
- Naor, D.; Nedvetzki, S.; Golan, I.; Melnik, L.; Faitelson, Y. CD44 in Cancer. Crit. Rev. Clin. Lab. Sci. 2008, 39, 527–579. [Google Scholar] [CrossRef] [PubMed]
- Murga-Zamalloa, C.; Rolland, D.C.; Polk, A.; Wolfe, A.; Dewar, H.; Chowdhury, P.; Önder, Ö.; Dewar, R.; Brown, N.A.; Bailey, N.G.; et al. Colony-Stimulating Factor 1 Receptor (CSF1R) Activates AKT/mTOR Signaling and Promotes T-Cell Lymphoma Viability. Clin. Cancer Res. 2019, 26, 690–703. [Google Scholar] [CrossRef] [Green Version]
- Chatterjee, S.; Azad, B.B.; Nimmagadda, S. The Intricate Role of CXCR4 in Cancer. In Advances in Cancer Research; Elsevier: Heidelberg, Germany, 2014; Volume 124, pp. 31–82. [Google Scholar]
- Sun, Y.; Zhao, C.; Ye, Y.; Wang, Z.; He, Y.; Li, Y.; Mao, H. High expression of fibronectin 1 indicates poor prognosis in gastric cancer. Oncol. Lett. 2019, 19, 93–102. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.J.; Choi, S.K.; Hong, S.H.; Eun, J.W.; Nam, S.W.; Han, J.-W.; You, J.S. Oncogenic IL7R is downregulated by histone deacetylase inhibitor in esophageal squamous cell carcinoma via modulation of acetylated FOXO1. Int. J. Oncol. 2018, 53, 395–403. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.-Y.; Zheng, C.; Xia, E.-J.; Quan, R.-D.; Hu, J.; Zheng, C.; Hao, R.-T. Lysophosphatidic Acid Receptor 5 (LPAR5) Plays a Significance Role in Papillary Thyroid Cancer via Phosphatidylinositol 3-Kinase/Akt/Mammalian Target of Rapamycin (mTOR) Pathway. Med. Sci. Monit. 2020, 26, e919820-1. [Google Scholar] [CrossRef]
- Juuti, A.; Lundin, J.; Nordling, S.; Louhimo, J.; Haglund, C. Epithelial MMP-2 Expression Correlates with Worse Prognosis in Pancreatic Cancer. Oncology 2006, 71, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.-J.; Wu, D.-W.; Wang, Y.-C.; Chen, C.-Y.; Lee, H. PAK1 confers chemoresistance and poor outcome in non-small cell lung cancer via β-catenin-mediated stemness. Sci. Rep. 2016, 6, 34933. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Cheng, G.; Song, Z.; Xu, T.; Ruan, H.; Cao, Q.; Wang, K.; Bao, L.; Liu, J.; Zhou, L.; et al. RAC2 acts as a prognostic biomarker and promotes the progression of clear cell renal cell carcinoma. Int. J. Oncol. 2019, 55, 645–656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urban-Wojciuk, Z.; Khan, M.M.; Oyler, B.L.; Fåhraeus, R.; Marek-Trzonkowska, N.; Nita-Lazar, A.; Hupp, T.; Goodlett, D.R. The Role of TLRs in Anti-cancer Immunity and Tumor Rejection. Front. Immunol. 2019, 10, 2388. [Google Scholar] [CrossRef] [PubMed]
- Papalexi, E.; Satija, R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat. Rev. Immunol. 2018, 18, 35–45. [Google Scholar] [CrossRef] [PubMed]

| Sr. No. | SRA IDs | Good Reads % | Alignment Rate |
|---|---|---|---|
| 1 | SRR8581331 | 98.79% | 98.10% |
| 2 | SRR8581332 | 99.70% | 98.63% |
| 3 | SRR8581333 | 99.60% | 98.54% |
| 4 | SRR8581334 | 99.71% | 98.46% |
| 5 | SRR8581335 | 99.68% | 98.39% |
| 6 | SRR8581336 | 99.59% | 98.53% |
| 7 | SRR8581337 | 99.71% | 98.45% |
| 8 | SRR8581338 | 99.71% | 98.45% |
| 9 | SRR8581339 | 99.66% | 98.56% |
| 10 | Average all sample | 99.57% | 98.45% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jaiswal, V.; Cho, Y.-I.; Lee, H.-J. Preliminary Study to Explore the Immune-Enhancement Mechanism of Platycodon grandiflorus Extract through Comparative Transcriptome Analysis. Appl. Sci. 2021, 11, 226. https://doi.org/10.3390/app11010226
Jaiswal V, Cho Y-I, Lee H-J. Preliminary Study to Explore the Immune-Enhancement Mechanism of Platycodon grandiflorus Extract through Comparative Transcriptome Analysis. Applied Sciences. 2021; 11(1):226. https://doi.org/10.3390/app11010226
Chicago/Turabian StyleJaiswal, Varun, Yeong-Im Cho, and Hae-Jeung Lee. 2021. "Preliminary Study to Explore the Immune-Enhancement Mechanism of Platycodon grandiflorus Extract through Comparative Transcriptome Analysis" Applied Sciences 11, no. 1: 226. https://doi.org/10.3390/app11010226
APA StyleJaiswal, V., Cho, Y.-I., & Lee, H.-J. (2021). Preliminary Study to Explore the Immune-Enhancement Mechanism of Platycodon grandiflorus Extract through Comparative Transcriptome Analysis. Applied Sciences, 11(1), 226. https://doi.org/10.3390/app11010226






