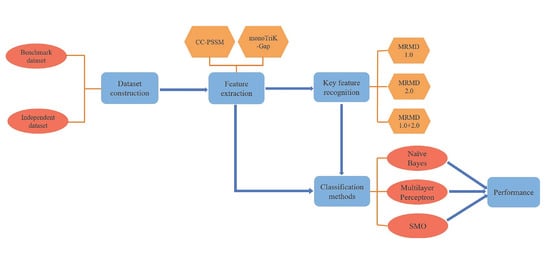
Accurate Prediction and Key Feature Recognition of Immunoglobulin
Abstract
Share and Cite
Gong, Y.; Liao, B.; Peng, D.; Zou, Q. Accurate Prediction and Key Feature Recognition of Immunoglobulin. Appl. Sci. 2021, 11, 6894. https://doi.org/10.3390/app11156894
Gong Y, Liao B, Peng D, Zou Q. Accurate Prediction and Key Feature Recognition of Immunoglobulin. Applied Sciences. 2021; 11(15):6894. https://doi.org/10.3390/app11156894
Chicago/Turabian StyleGong, Yuxin, Bo Liao, Dejun Peng, and Quan Zou. 2021. "Accurate Prediction and Key Feature Recognition of Immunoglobulin" Applied Sciences 11, no. 15: 6894. https://doi.org/10.3390/app11156894
APA StyleGong, Y., Liao, B., Peng, D., & Zou, Q. (2021). Accurate Prediction and Key Feature Recognition of Immunoglobulin. Applied Sciences, 11(15), 6894. https://doi.org/10.3390/app11156894