Abstract
Pelagic chlorophyll-a concentrations are key for evaluation of the environmental status and productivity of marine systems, and data can be provided by in situ measurements, remote sensing and modelling. However, modelling chlorophyll-a is not trivial due to its nonlinear dynamics and complexity. In this study, chlorophyll-a concentrations for the Helgoland Roads time series were modeled using a number of measured water and environmental parameters. We chose three common machine learning algorithms from the literature: the support vector machine regressor, neural networks multi-layer perceptron regressor and random forest regressor. Results showed that the support vector machine regressor slightly outperformed other models. The evaluation with a test dataset and verification with an independent validation dataset for chlorophyll-a concentrations showed a good generalization capacity, evaluated by the root mean squared errors of less than 1 µg L−1. Feature selection and engineering are important and improved the models significantly, as measured in performance, improving the adjusted R2 by a minimum of 48%. We tested SARIMA in comparison and found that the univariate nature of SARIMA does not allow for better results than the machine learning models. Additionally, the computer processing time needed was much higher (prohibitive) for SARIMA.
1. Introduction
Pelagic chlorophyll-a concentrations (chl-a) are a common indicator of primary production and key to evaluation of the health and productivity of marine and freshwater systems [1,2]. It is therefore of crucial importance to accurately measure/predict chlorophyll from proxy parameters in such systems [3]. Accelerated global warming is exacerbating climate change and unsettling ecosystems’ processes, while the impacts of this are directly affecting marine primary production and triggering an upwards transfer of effects that reach humans. Thus, the importance of modelling chlorophyll is emphasized in environments undergoing change resulting from global warming [4].
Prediction of chlorophyll-a time series data is a challenge due to their complexity and nonlinearity, and indeed, conventional approaches show limitations with prediction of unobserved data [5,6]. To date, all conventional approaches, including factors based on single measurements, are limited with regard to prediction accuracy of chlorophyll-a concentrations [7]. A few previous studies have tried to implement various machine learning techniques to predict chlorophyll concentrations, mainly in fresh water systems, with a few in marine regions [8,9,10,11].
Machine learning (ML) techniques constitute a set of tools belonging to the fields of computer science and artificial intelligence. The versatility of these techniques allow the successful application in many fields of science and to a great variety of problems. The focus is often placed on tackling pattern recognition problems and on the construction of predictive models to make data-driven decisions [12]. According to [13], the general benefits of ML algorithms for time series prediction over classical methods include the ability of supporting noisy features, noise and complexity in the relationships between variables and in the handling of irrelevant features.
State-of-the-art ML algorithms for time series regression include random forest regressor (RF), support vector machine regressor (SVR) and neural networks multi-layer perceptron regressor (MLP). All of these have been used to some degree in the literature for the prediction of chlorophyll-a concentrations in aquatic systems, and have achieved significantly accurate results in both error and goodness of fit metrics [3,11,14]. These are studies based in chl-a time series either with short length and daily frequency or long-term, low frequency sampling time series, using different ML methods to best predict chl-a behavior. The features applied as predictors in these studies are limited to just a few, but it must be considered that the dynamics in lacustrine systems are distinct from those presented in marine systems. Here we extend these ideas and test these methods on a good quality long-term time series, the Helgoland Roads time series, evaluating the prediction using unseen data. With the purpose to compare ML methods with a classical statistical regression model, we included an improved autoregressive integrated moving average (ARIMA) model, called seasonal ARIMA (SARIMA), which includes seasonal parameters to support data with a seasonal component [15].
The objective of this work is to evaluate the accuracy of machine learning algorithms for the estimation of chlorophyll-a concentration, using in situ high resolution long-term datasets. We (1) assess three ML algorithms—random forest, support vector regressor and neural networks multi-layer perceptron regressor—for chlorophyll-a concentration estimation; (2) examine the importance of feature selection and engineering in the different models; (3) compare with, and evaluate, a univariate SARIMA classical regression model.
2. Materials and Methods
All the ML models used in this study were implemented applying the “Scikit-Learn package”, which is an open-source Python module project that integrates a wide range of common ML algorithms [16,17], while the SARIMA model was implemented with the statsmodels package [18]. The preprocessing was also implemented in the Python environment, using the well-known packages Pandas, NumPy and SciPy [19].
2.1. Datasets
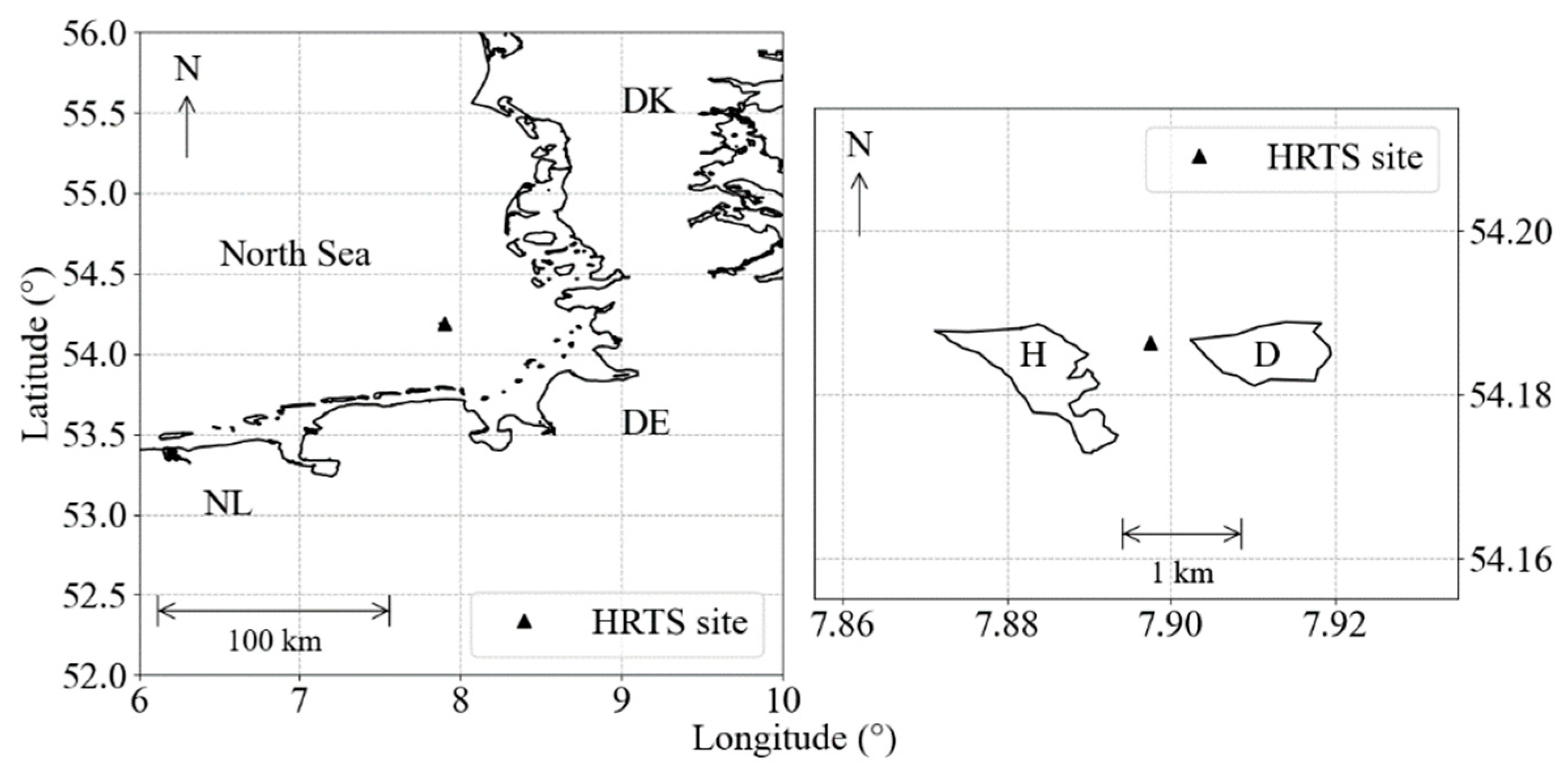
The Helgoland Roads is a long-term pelagic monitoring site (54°11.3′ N, 7°54.0′ E) about 60 km off the German coast and represents a marine transition zone between coastal waters and open sea (Figure 1) [20]. Since 1962, surface water samples have been collected on working days, taken with a bucket lowered from a research vessel. Secchi depth and water temperature (SST) are measured in situ and the water samples analyzed in the laboratory for nutrients (nitrate, phosphate and silicate) and salinity. Chlorophyll-a concentration measurements were started at the end of 2001, acquired in laboratory by FluoroProbe (bbe Moldaenke GmbH, Kiel, Germany) [21] and, since, 2004 have been complemented with high-performance liquid chromatography analysis (HPLC) [22,23].
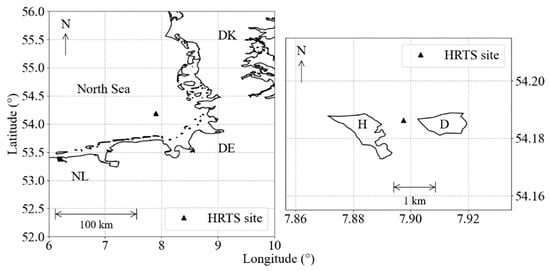
Figure 1.
Helgoland Roads monitoring site position (black triangle) in the German Bight, between the Helgoland (H) and Dune (D) islands.
Sunshine duration, wind speed and direction [24,25,26], North Atlantic Oscillation (NAO) daily index (NOAA ESRL Physical Sciences Laboratory, Boulder, CO, USA, 2020) and zooplankton abundance [27], were added to the Helgoland Roads parameter matrix for this work (Table 1). As indicated in the literature [28,29,30], and also from working experience, the included parameters are environmental variables which determine algal verdure and, thus, modulate chlorophyll-a concentrations in marine systems.

Table 1.
Statistical description of parameters used as determinants to predict chlorophyll-a concentration (target) after linear interpolation (std, min and max are standard deviation, minimum and maximum values, respectively).
2.2. Data Preprocessing
The raw data of Helgoland Roads are characterized by long-term measurements on work-daily frequency, with missing values during weekends and extreme bad weather days. When merged with date of other features such as zooplankton abundance, it ends with approximately 40% of missing data in the time series. To fill the missing data and creating a regular sampled daily time series, a number of imputation methods were tested in sunlight duration, a feature added to the Helgoland Roads from an external source, with no missing values. After creating a synthetic missing values dataset with sunlight duration, we calculated root mean square error (RMSE) and coefficient of determination (R2) between the original and interpolated data. Minimum changes in frequency distribution between missing data and interpolated variables, lowest RMSE and highest R2, were the basis for the decision to use a linear interpolation, supported by [30]. After the interpolation, we have daily datasets of parameters in Table 1 comprising approximately 13 years, from 2 November 2001 to 22 April 2015, and presented in Supplementary Materials, Figure S1.
In this study, to validate the performance of the ML models, the dataset was split in 80% (n = 3940) for model training, and 20% (n = 980) for model testing, so we could investigate the model generalization ability [31]. To eliminate the dimensional differences of the data and improve the prediction ability of the models, we used the StandardScaler method from the Scikit-Learn package, which standardizes features by removing the mean and scaling to unit variance.
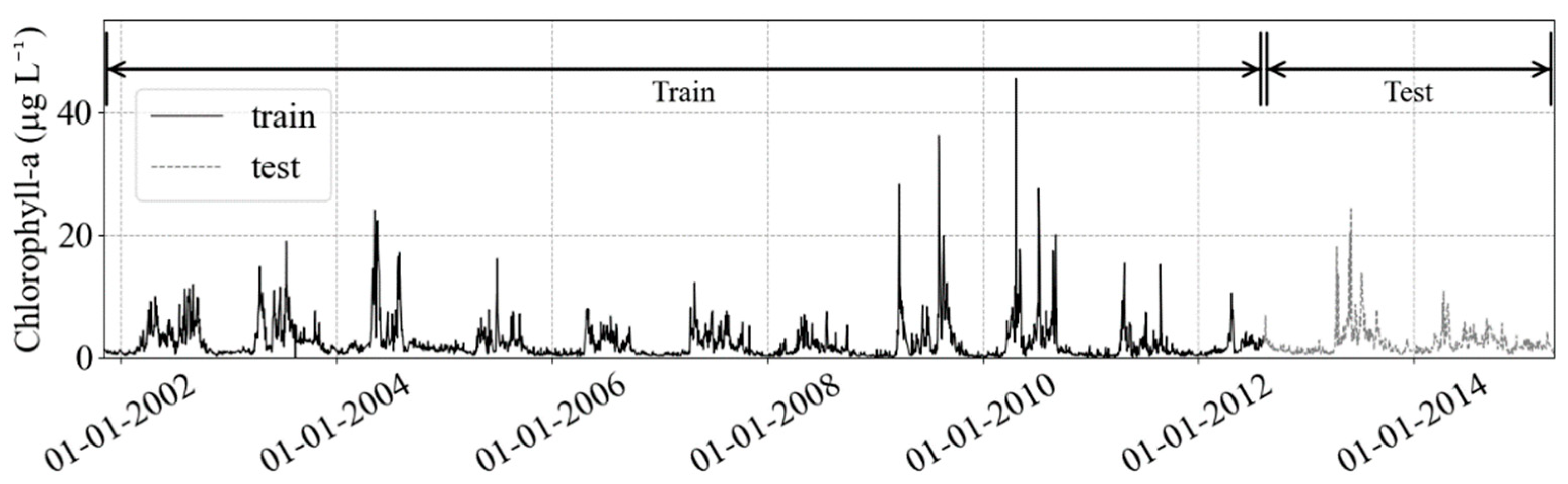
The training dataset, the sample of data used to fit the model, dates from 2 November 2001 to 15 August 2012 (~11 years), while the test set is from 16 August 2012 to 22 April 2015 (~2.5 years) and it is used for model evaluation (Figure 2). For independent validation, we used a linear interpolated time series of HPLC estimated chlorophyll data (5 May 2015 to 27 November 2018, n = 348).
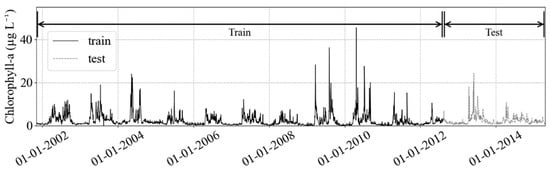
Figure 2.
The train and test partition in chlorophyll-a concentration target (black solid and gray solid lines, respectively), and the HPLC chl-a validation dataset (black dashed). After the split, the testing dataset will remain untouched, to guarantee no leakage of information to the training step. The validation dataset is the independent validation.
2.3. Feature Engineering and Selection
The Pearson correlation coefficients were calculated to investigate linear relationships between chlorophyll-a concentration and the other variables (Table 2). All correlation coefficients were lower than 0.5, indicating no strong linear correlation between chlorophyll and any other variable.

Table 2.
Pearson correlation among predictors and the target chlorophyll-a concentration.
Prediction is a major task of time series data mining, which uses known historical values to estimate future values, and feature selection and engineering is essential and crucial for accurate predictions [32]. To seek improvement, 15 days lagged predictors were generated, totalizing 211 features [33]. The choice of lags was based in a two-week period where all the predictors supposedly influence chlorophyll-a concentration, including chl-a past values, i.e., the lagged target values were used as predictors (t − 1, …, t − n; with t as the current time and n = 15). As there are significant seasonal differences, e.g., summer and winter nutrients uptake, the definition of two weeks seemed reasonable for this work to input information, considering that the machine learning algorithms are data-driven and they are not mechanistic models [34]. Additionally, date features were generated, namely, “year” and “day of year” from 1 to 365 or 366. The cyclic variables “day of year” and “wind direction” were transformed with
to ensure that the last day of a year was understood to be in sequence with the first day of the next year and 0° degree in direction was equal to 360° [35].
sin [2π (day of year)/(number of days in year)]
cos [2π (day of year)/(number of days in year)]
sin [2π (wind direction (°))/(360)]
cos [2π (wind direction (°))/(360)]
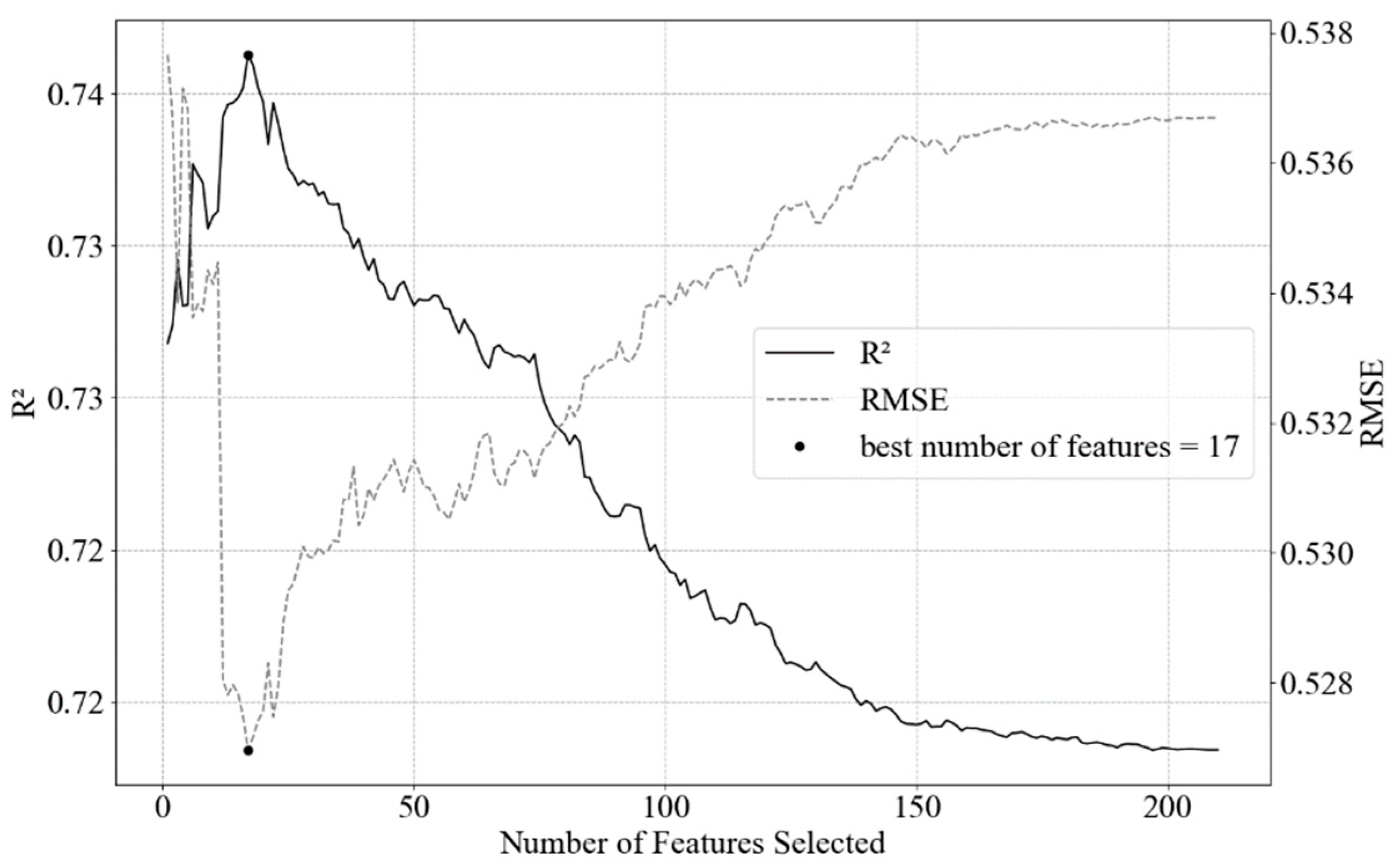
A large number of features in the dataset drastically affects both the training time as well as the accuracy of machine learning models. One means to limit model complexity from multiple variables is to reduce the model by selectively eliminating predictors. Feature selection procedure was conducted applying a combination of Recursive Feature Elimination. We used Scikit-Learn module Recursive Feature Elimination with cross-validation (Scikit-Learn feature.selection RFECV module) and Ridge estimator, to estimate the best number of features balanced with accuracy (Figure 3). After the best number of features were defined with the Ridge cross-validation method, we applied Recursive Feature Elimination (Scikit-Learn feature.selection RFE module) with SVR linear estimator, this way selecting the 17 best parameters to model chl-a in a robust manner (Table 3) [36].
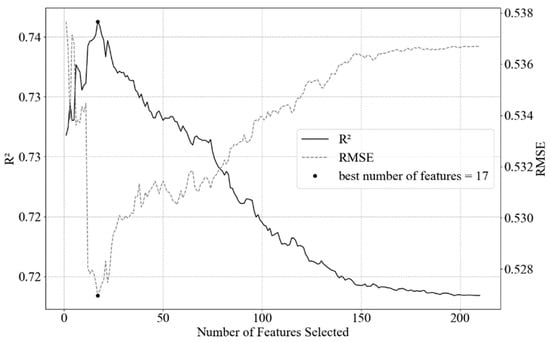
Figure 3.
Result of RFECV with Ridge estimator. The black dot represents the maximum value of 17 selected features (predictors) to reach the highest explained variance. After the maximum value, there is an exponential decay/increase in the R2/RMSE. RMSE unit is µg L−1.

Table 3.
Chosen features after Feature Selection process. The negative numbers represent the number of lags in the original features (t − 1, …, t − 15).
2.4. Model Selection and Hyperparameter Tuning
The algorithms evaluated in this study are random forest regressor (RF) [37], support vector machine regressor (SVR) [38] and multi-layer perceptron regressor neutral network (MLP) [39,40]. These were chosen to be widely used and to present available information that allows the easy application in any level of knowledge concerning ML. Compared with deep learning approaches, traditional machine learning does not need large amounts of data to train and the computer processing can be performed in low-end machines without a GPU (Graphics Processing Unit) [41].
SVR is a kernel-based nonlinear regression method. It transforms the original input data space into a high-dimensional input space (hyperplanes) and performs linear regression in the high-dimensional space by defining a maximum margin separator, which minimizes expected generalization error instead of the prediction error in the training dataset. The kernel functions, which take as input the dot products of pairs of input points, allows the SVR to map the inputs efficiently compared to calculating the corresponding points of each input in the high-dimensional space. Basically, SVR finds hyperplanes that minimize the errors and maximize the margins of continuous data [6].
RF is a machine learning technique that utilizes an ensemble of decision trees for regression tasks. It randomly takes subsets of the data and input variables, and the results of all trees are averaged to achieve a better result than individual trees. The use of random samples of the training data for multiple decision trees reduces overfitting compared to using the entire training set with a single decision tree [42].
MLP is an artificial neural network and it consists of connected nodes, resembling the neurons in a biological brain. It consists of at least three layers of nodes: the input layer, hidden layer and output layer. Excluding the input layer nodes, each node receives inputs from the other nodes, and the outputs are calculated using a nonlinear activation function. The learning process for MLP involves continually adjusting weights in the network to minimize the error rate using backpropagation. Backpropagation computes the gradient of the loss function with respect to the weights and updates the weights in the network using methods such as stochastic gradient descent [42].
Depending upon the study cases, different ML algorithms usually require some adjustments. These are often crucial for the development of a successful application. Each ML algorithm has parameters, so-called hyperparameters, which define the setup of the machine to modelling the target function. For each model, a search range of hyperparameters was tested. In cases where a value was selected at the edge of the search range, a new cross-validation was conducted including more values.
All hyperparameter tuning of the models (Table 4) is based on GridSearchCV in the Scikit-Learn package, which can evaluate all possible given combinations of hyperparameter values using 10-fold cross-validation. This procedure determines the best combination of hyperparameters of the model that gives the best accuracy, in terms of coefficient of determination (R2).

Table 4.
Hyperparameters tested in GridSearchCV and those applied to each ML algorithms.
Cross-validation is a model validation technique for obtaining reliable and stable models. The use of multiple models in the evaluation removes possible biases of some models with some data sets. We used the training dataset to search for the best parameters, and reported the prediction performances on the test dataset using these parameters [43]. The mentioned grid search was performed independently for each model on the training subset.
R2, adjusted coefficient of determination (adj R2) and RMSE were the metrics used in this work to evaluate the predictions. The use of adj R2 in multiple regression is important because it increases only when new independent variables that increase the explanatory power of the regression equation are added; this makes it a useful measure of how well a multiple regression equation fits the sample data. A linear base model, available in Scikit-Learn, was used to observe the improvements using the more sophisticated algorithms.
2.5. SARIMA Model
For the SARIMA model, the univariate chl-a data was used, while maintaining the partitions in the training and test dataset. To test stationarity, the Augmented Dickey–Fuller test (ADF) was applied indicating significant stationarity (p < 0.05) in the train and test datasets. To fill the model (p, d, q) × (P, D, Q)365, where 365 represents the seasonality, the best autoregressive (p, P) and moving average (q, Q) parameters were selected using an iterative method in the train dataset. The parameters ranged from 0 to 4 in the nonseasonal parameters (p, q) and 0 to 2 in the seasonal parameters (P, Q), selecting the combination with lowest Akaike information criterion (AIC). The difference order parameters d and D were 0, due to the stationarity results of the ADF test. The best parameters selected using the training dataset were (4, 0, 1) × (2, 0, 1)365, and this SARIMA model was used to fit the test dataset.
3. Results
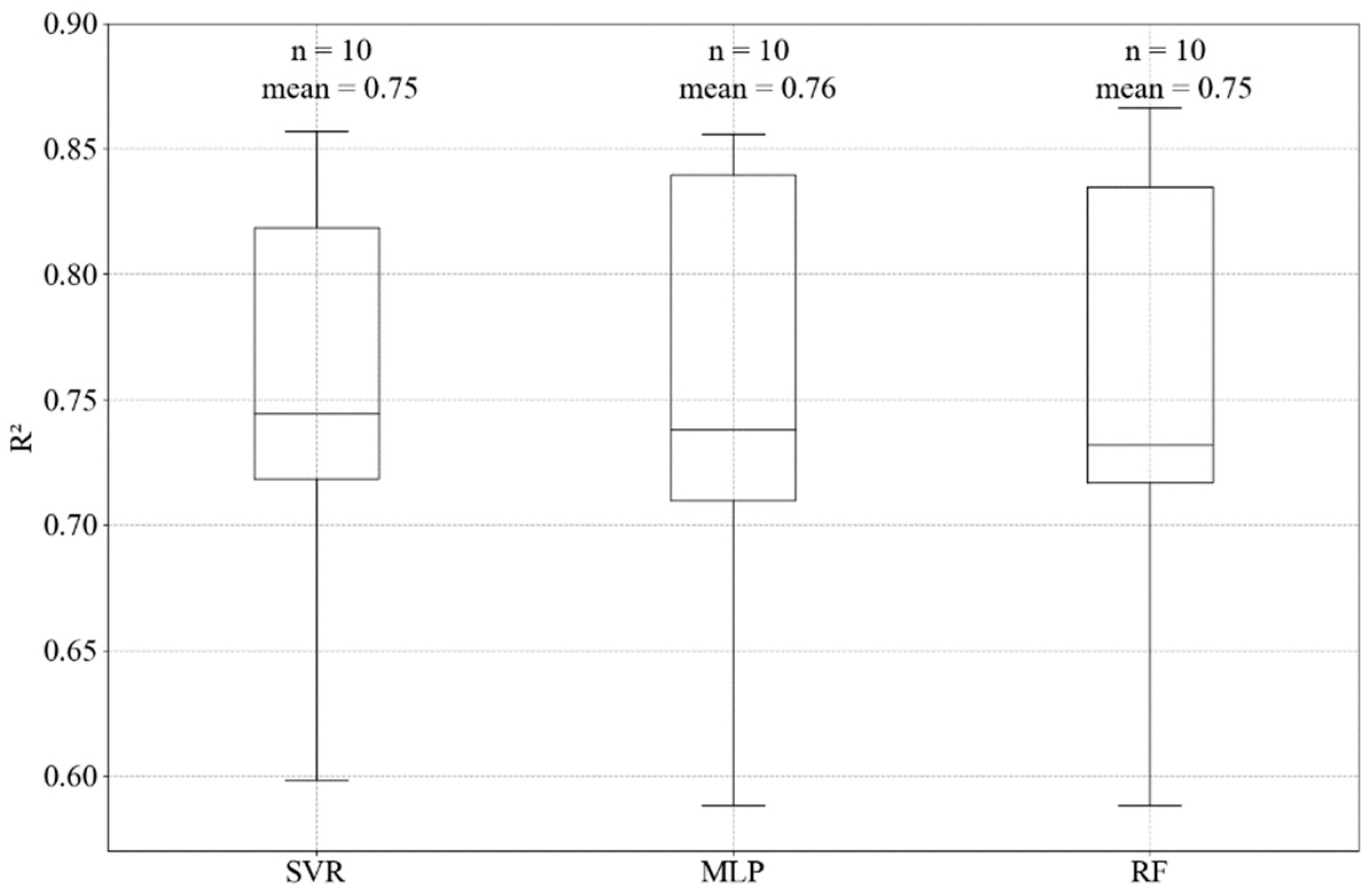
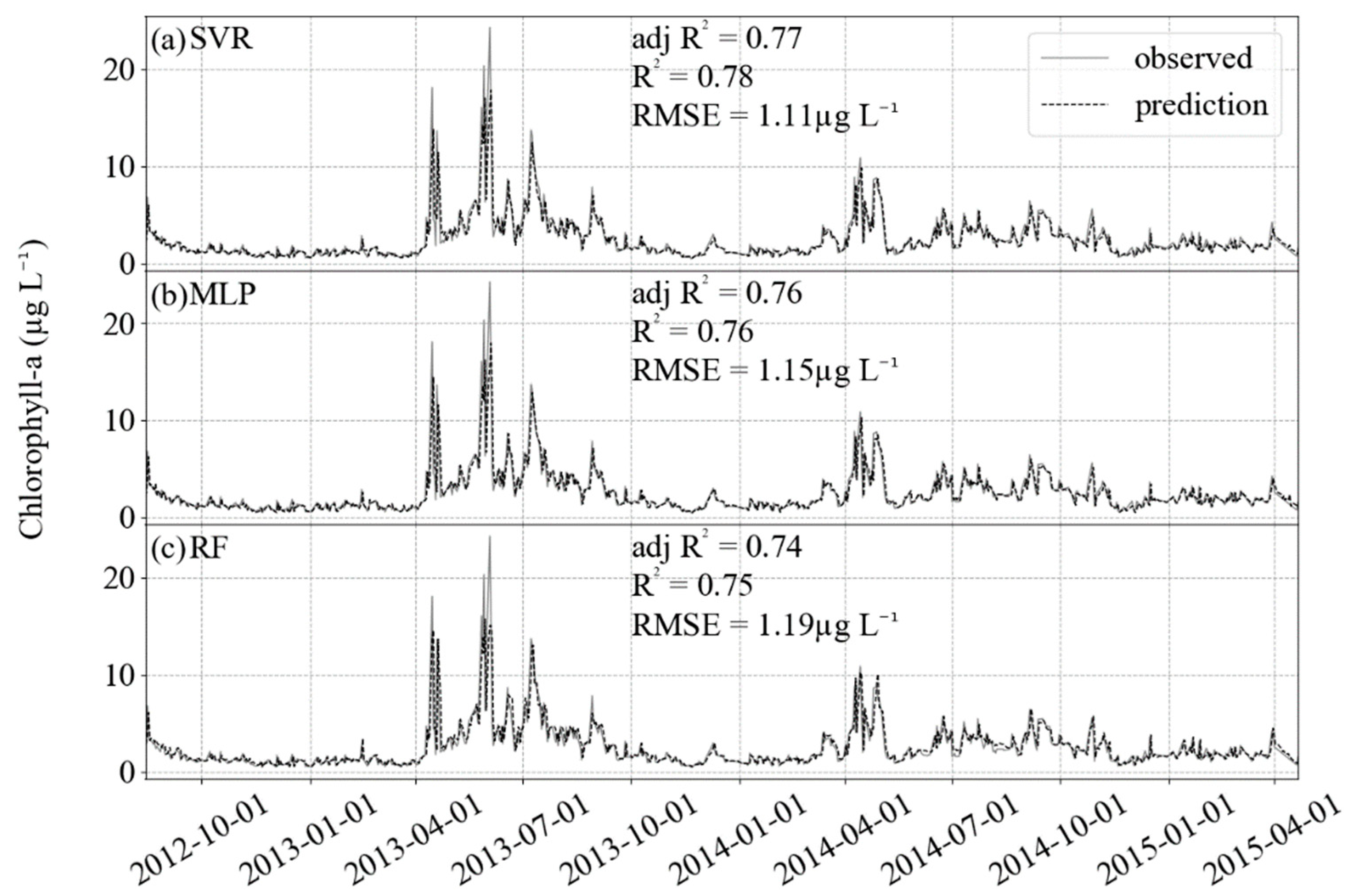
For this study, the best R2, adj R2 and RMSE achieved for predicting chlorophyll-a using support vector machine regressor, random forest regressor, and neural network multi-layer perceptron regressor are presented in Table 5. In a combination of hyperparameters tuning and feature selection, the models showed improvement compared with the default models (no feature selection, no tuning) for the test datasets. Comparing the algorithms, SVR reached the best R2 (0.78) and RMSE (1.113 µg L−1), however, these were only slightly better results (MLP = 0.76; 1.144 µg L−1 and RF = 0.75; 1.189 µg L−1). The algorithms presented good performances for the subsets of training dataset during the cross-validation step (Figure 4). In addition, the predicted values were close to the observed data (Figure 5). All the ML algorithms were better than the linear base model.

Table 5.
Comparison of nonoptimized (default) and optimized model performances for predicting chlorophyll-a concentration during training (train) and testing (test) steps. The linear model serves as a base model.
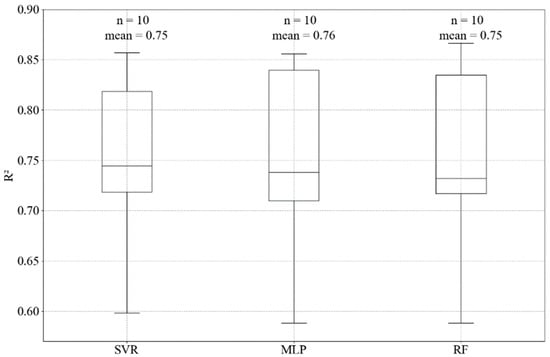
Figure 4.
Boxplot of accuracy in the 10-fold cross-validation training step for the SVR, MLP and RF models, showing the mean and the number of folds (n) or subsets in the training data used to define the best hyperparameters.
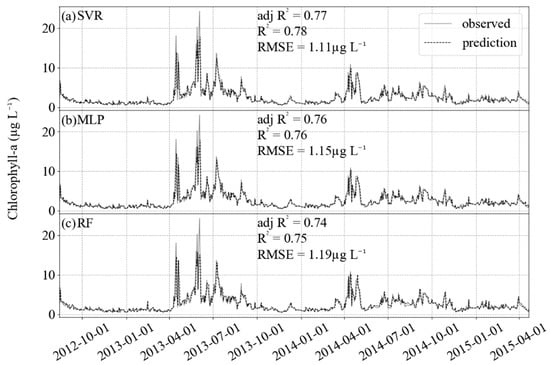
Figure 5.
Results of prediction (black dashed) and comparison with the observed test dataset (gray solid). For the three algorithms, R2 is higher than 0.7 and RMSE lower than 1.2 µg L−1. (a) SVR, (b) MLP and (c) RF.
The algorithms gave a good performance for the training dataset and allowed a good generalization for the test dataset, as it can be seen from how close the predicted values are from those observed in Figure 5. Using all of the 211 features and the default hyperparameters, the results in the test data were not as good as those from the optimized models (Table 5), mainly due to overfitting, when the models are more complex than necessary and the fitting in the training dataset is affected by noise [44].
Considering the features used as inputs in each of the algorithms, the Recursive Feature Elimination was implemented by combining Ridge and SVR linear estimators and selecting a maximum number of 17 predictors. This generated the following result: (‘SD, ‘SST’, ‘Salinity’, ‘SD_-1’, ‘SST_-1’, ‘SST_-2’, ‘SST_-9’, ‘SST_-12’, ‘SST_-13’, ‘SST_-14’, ‘SST_-15’, ‘Salinity_-1’, ‘Chl_-1’, ‘Chl_-4’, ‘Chl_-5’, ‘Chl_-7’, ‘Chl_-8’), with the negative numbers in the codes (Table 2) representing the applied lag in days. The adj R2 results, which are sensitive to the number of used predictors, showed improvement from 0.02 to 0.76 for MLP, while for SVR the result improved from 0.63 to 0.77 and from 0.15 to 0.74 for RF in the test dataset.
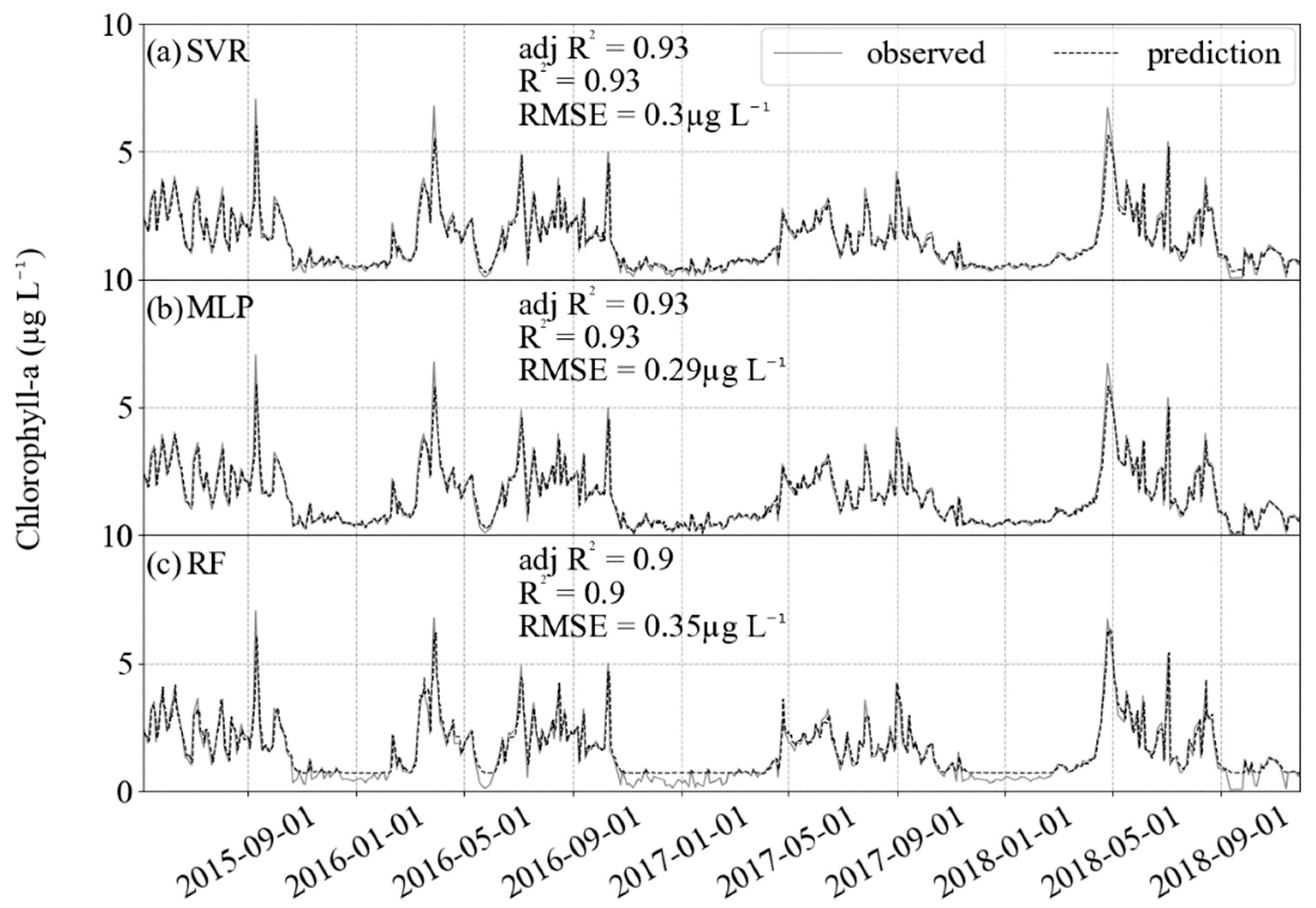
For the independent validation, a chl-a dataset acquired by HPLC, the predictions had better RMSE and R2 than the test datasets (Figure 6). Again, the higher values had limitations in prediction, but the lower variance compared with the training and testing datasets allowed for better evaluation indicators, with RMSE for all algorithms in the order of 0.3 µg L−1 and R2 reaching approximately 0.90.
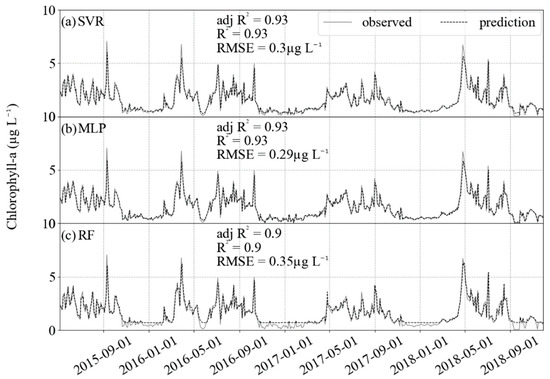
Figure 6.
Results of prediction (black dashed) and comparison with the validation dataset (gray solid). For the three algorithms, R2 is approximately 0.9 and RMSE lower than 0.3 µg L−1. (a) SVR, (b) MLP and (c) RF.
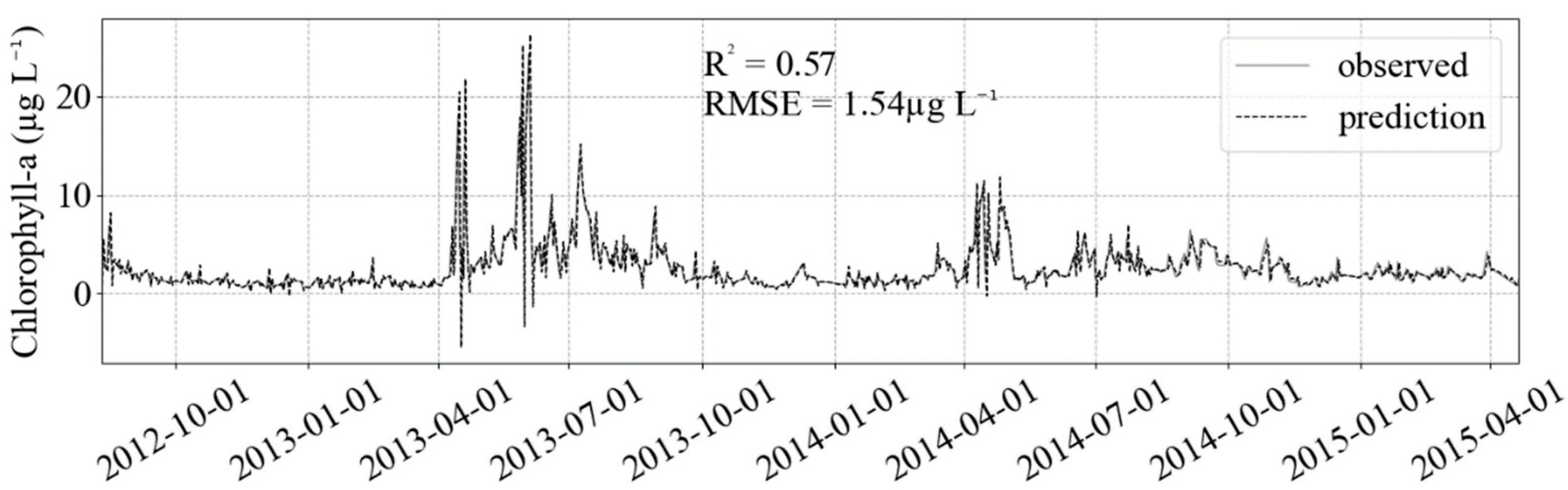
The iterative SARIMA parameters selection uses much more computer processing time compared with the GridSearchCV method in machine learning. The latter is a scale of seconds to minutes while the former hours to days. It took around two weeks to select the best p, q, P and Q parameters in the daily data considering a yearly seasonality. Fitting the test dataset with the SARIMA model gave the worst results when compared with the ML models (Figure 7).
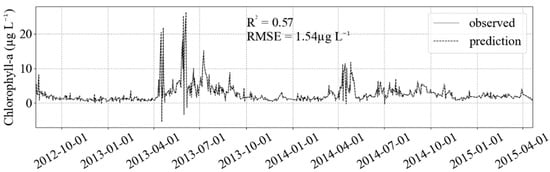
Figure 7.
Result of SARIMA fit (black dashed) in the test dataset (gray solid). The better fit in extreme values is counter-balanced by the estimation of negative values, decreasing/increasing R2/RMSE compared to the ML models results.
4. Discussion
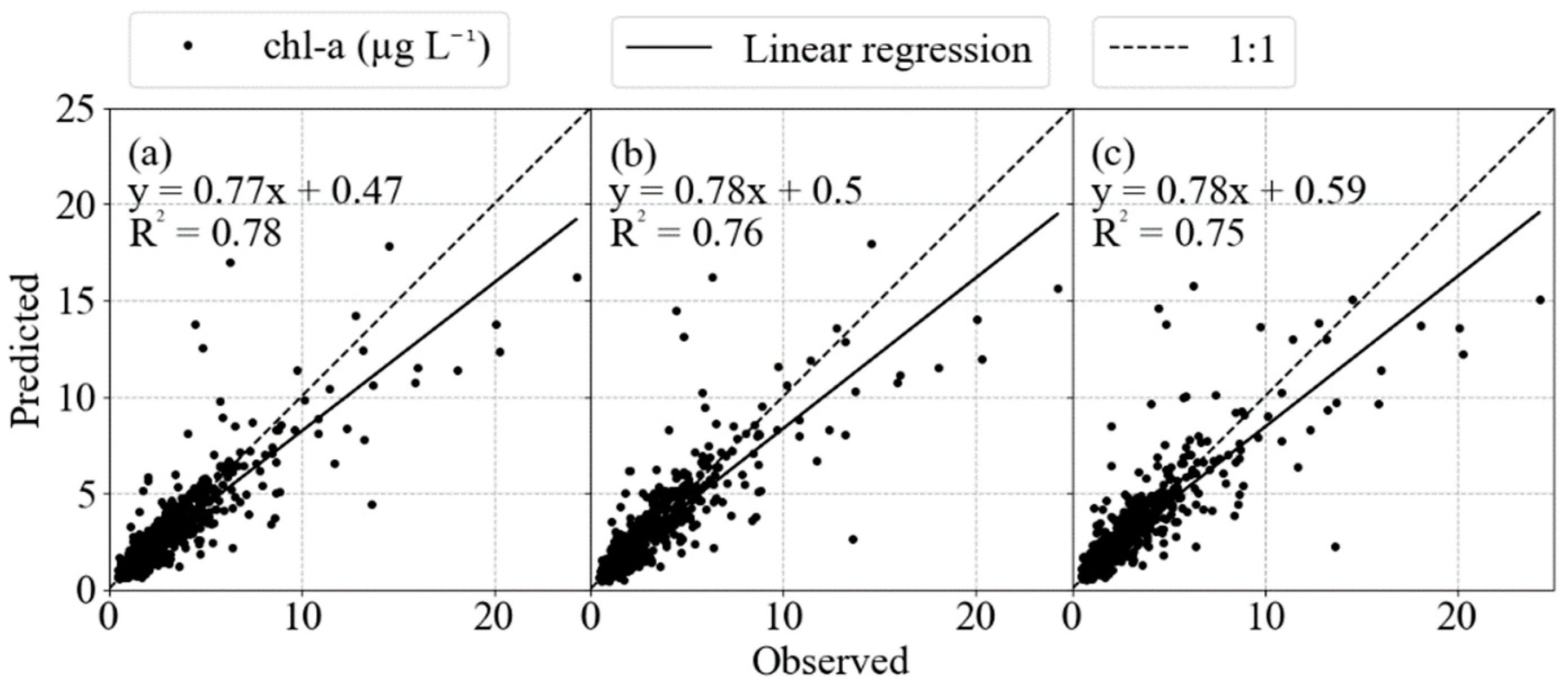
Machine learning analysis was conducted on the Helgoland Roads time series to develop the best fit of chlorophyll-a concentrations over time using different parameters and their lagged correlates. For the three algorithms implemented, the model results were virtually equal in the evaluation metrics, presenting similar results in prediction, with slightly better values for the model SVR. For the time predictions, each of the three models’ performances are acceptable with high R2 values greater than 0.70 and RMSE lower than 1.5 μg L−1, ~40% smaller than the chlorophyll-a concentration standard deviation of 2.9 μg L−1. However, all of the algorithms were unable to predict extreme values (Figure 8). It was expected that a certain degree of decrease in accuracy would be incurred because of the difficulty in capturing and reproducing these extreme peaks [45]. One hypothesis that would explain the underestimation of extreme values is the absence of predictive features, e.g., hydrodynamics can result in the transport of chlorophyll from other areas as an input event, even though salinity and wind parameters are reliable indicatives for current and wave dynamics in the German Bight [46]. As these events do not present as a temporal pattern, the ML models do not recognize the influence on the target.
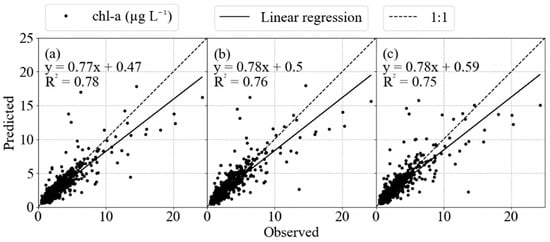
Figure 8.
Cross-plots of the modeled and observed chlorophyll values in (a) SVR, (b) MLP and (c) RF. It is possible to notice the deviation in extreme values, showing the limitation of the ML models in deal with these data values.
Because each algorithm is based on different algebraic assumptions and procedures, they can result in different predictions. Between SVR and MLP, [14] points to differences in the nonlinear equalization performance and the structural risk minimization principle of SVR being more effective than the empirical risk minimization principle of neural networks in terms of minimizing error. According to [47], in MLP the method for determining global solutions is difficult to converge because of its inherent algorithm design and model parameters are more complex than SVR, whereas the SVR has ready access to global optimal solutions, obtained by solving a linearly constrained quadratic programming problem [14]. Between SVR and RF, as we saw, the linear base model gave good results. There is the possibility of a linear dependency that is better captured by SVR, probably a result from the linear interpolation in the preprocessing step of this study.
The feature selection and tuning of hyperparameters was extremely important and improved the results substantially. This was noticeable in the adj R2 results for default and optimized models. Analyzing the 17 features used in SVR and described in the results section, the algorithm considered SST, lagged SST, lagged chlorophyll, salinity and Secchi depth to reach the best results presented in this work. It is important to point that ML is a data-driven approach, but it is possible to make inferences about the selected features. The number of selected features was a response of balancing bias and variance in the learning algorithms [48]. For this study, we noticed the choice of SST as an important feature, probably representing the seasonal patterns in the chlorophyll target.
Better R2, adj R2 and RMSE results in the independent validation dataset are possibly due to less variability and absence of extreme values, and shows the good generalization that the ML models are capable of. All of the good results, for both the test and independent validation data, show the better prediction power of the three ML algorithms evaluated in this study. Comparing with the classical SARIMA model, the univariate and linear background did not achieve the results needed for it to outperform the ML models. Compared with the ML literature, studies such as [3] and [11] achieved results of R2 ranging from 0.50 to 0.80, analyzing shorter time series of chl-a in lakes. The authors of [49] predicted variations of chlorophyll-a in different sites of the North Sea using generalized additive models (GAM) and the R2 results ranged from 0.15 to 0.63. In [28], using GAM to predict chl-a in a spatial approach for the North Atlantic, got the best result for R2 at 0.83. All of these values show how variable different methods’ performances in predicting chlorophyll can be, not necessarily meaning one method is better than the other, but more adaptive. ML models proved their generalization capacity and high accuracy.
5. Conclusions
In this work, we evaluated three machine learning algorithms in a regression task. Support vector regressor presented a slightly better performance, with the advantage that it used less computational time, and generated chlorophyll concentration predictions with 0.78 correlation to the observed data, in comparison to 0.76 and 0.75 for MLP and RF, respectively. Moreover, the root mean square error was approximately 1.1 µg L−1 for the test dataset and less than one for the independent validation data, which is approximately 38% percent smaller than the standard deviation of 2.9 µg L−1. This study demonstrates the ability of machine learning models to use environmental in situ time series to predict the chlorophyll concentration with significant accuracy (R2), higher than 70%, and the importance of tuning hyperparameters and defining the best predictors (feature selection). Most chlorophyll-a prediction studies are conducted in fresh water environments or using satellite data and limited time series, so this work can be considered a step toward the use of machine learning algorithms in marine areas based on long-term time series. Being aware of the limitations presented in this study, in future works it would be interesting to work with irregular sampled time series, improve the method for feature selection, ensemble results of different ML and classical statistical models, and evaluate the forecasting power of these models in the short and long term. Besides, the use of deep learning approaches has become more and more common, and in many cases, they are outperforming the traditional ML algorithms, with the cost of higher computer processing times.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/app11167208/s1, Figure S1: Time series of parameters used to predict chlorophyll-a concentration: (a) Secchi disk depth, in meters (m); (b) Sea Surface Temperature, in degrees Celsius (°C); (c) Salinity (−); (d) Silicate (µmol L−1); (e) Phosphate (µmol L−1); (f) Nitrate (µmol L−1); (g) Sunlight duration, in hours (h); (h) NAO index (−); (i) Wind Direction, in degrees (°); (j) Wind Speed, in meters per second (m s−1); and (l) Total zooplankton abundance, individuals per cubic meter (ind. m−3).
Author Contributions
Conceptualization, F.d.L.L.d.A.; methodology, F.d.L.L.d.A.; formal analysis, F.d.L.L.d.A.; data curation, J.R.; writing—original draft preparation, F.d.L.L.d.A.; writing—review and editing, G.L., J.R. and K.H.W.; visualization, F.d.L.L.d.A.; supervision, G.L. and K.H.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from Karen Helen Wiltshire (karen.wiltshire@awi.de). The data are not publicly available, for our archives and financial agencies we have to document to whom and which time series data have been made available.
Acknowledgments
We acknowledge the present and past crews of the research vessels Aade and Ellenbogen of the Biologische Anstalt Helgoland (BAH) for their unfailing provision of samples. We also thank Kristine Carstens, Silvia Peters, Ursula Ecker and all colleagues of time series group and those who were instrumental in the analysis of samples during the last decades. We thank the NOAA Physical Sciences Laboratory and Deutsche Wetterdienst for providing climate and meteorological data. We acknowledge support by the Open Access Publication Funds of Alfred-Wegener-Institut Helmholtz-Zentrum für Polar- und Meeresforschung. We thank the Editor and two anonymous reviewers for their careful evaluation of our manuscript, the positive feedback, and their comments that helped to further improve the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Huot, Y.; Babin, M.; Bruyant, F.; Grob, C.; Twardowski, M.S.; Claustre, H. Does chlorophyll a provide the best index of phytoplankton biomass for primary productivity studies? Biogeosciences 2007, 4, 853–868. [Google Scholar] [CrossRef] [Green Version]
- Terauchi, G.; Tsujimoto, R.; Ishizaka, J.; Nakata, H. Preliminary assessment of eutrophication by remotely sensed chlorophyll-a in Toyama Bay, the Sea of Japan. J. Oceanogr. 2014, 70, 175–184. [Google Scholar] [CrossRef]
- Luo, W.; Zhu, S.; Wu, S.; Dai, J. Comparing artificial intelligence techniques for chlorophyll-a prediction in US lakes. Environ. Sci. Pollut. Res. 2019, 26, 30524–30532. [Google Scholar] [CrossRef]
- Botkin, D.B.; Saxe, H.; Araujo, M.B.; Betts, R.; Bradshaw, R.H.; Cedhagen, T.; Chesson, P.; Dawson, T.P.; Etterson, J.R.; Faith, D.P. Forecasting the effects of global warming on biodiversity. BioScience 2007, 57, 227–236. [Google Scholar] [CrossRef]
- Shamshirband, S.; Jafari Nodoushan, E.; Adolf, J.E.; Abdul Manaf, A.; Mosavi, A.; Chau, K.W. Ensemble models with uncertainty analysis for multi-day ahead forecasting of chlorophyll a concentration in coastal waters. Eng. Appl. Comput. Fluid Mech. 2019, 13, 91–101. [Google Scholar] [CrossRef] [Green Version]
- Shin, Y.; Kim, T.; Hong, S.; Lee, S.; Kim, T.; Park, M.S.; Park, J.; Heo, T.-Y. Prediction of Chlorophyll-a Concentrations in the Nakdong River Using Machine Learning Methods. Water 2020, 12, 1822. [Google Scholar] [CrossRef]
- Kwiatkowska, E.J.; Fargion, G.S. Application of machine-learning techniques toward the creation of a consistent and calibrated global chlorophyll concentration baseline dataset using remotely sensed ocean color data. IEEE Trans. Geosci. Remote Sens. 2003, 41, 2844–2860. [Google Scholar] [CrossRef]
- Cho, H.; Choi, U.J.; Park, H. Deep Learning Application to Time Series Prediction of Daily Chlorophyll-a Concentration. WIT Trans. Ecol. Environ. 2018, 215, 157–163. [Google Scholar] [CrossRef] [Green Version]
- Krasnopolsky, V.; Nadiga, S.; Mehra, A.; Bayler, E. Adjusting Neural Network to a Particular Problem: Neural Network-Based Empirical Biological Model for Chlorophyll Concentration in the Upper Ocean. Appl. Comput. Intell. Soft Comput. 2018, 2018, 7057363. [Google Scholar] [CrossRef]
- Keller, S.; Maier, P.M.; Riese, F.M.; Norra, S.; Holbach, A.; Börsig, N.; Wilhelms, A.; Moldaenke, C.; Zaake, A.; Hinz, S. Hyperspectral Data and Machine Learning for Estimating CDOM, Chlorophyll a, Diatoms, Green Algae and Turbidity. Int. J. Environ. Res. Public Health 2018, 15, 1881. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Feng, J.; Wang, Y. Chlorophyll a predictability and relative importance of factors governing lake phytoplankton at different timescales. Sci. Total Environ. 2019, 648, 472–480. [Google Scholar] [CrossRef] [PubMed]
- Lo, A.W.; Siah, K.W.; Wong, C.H. Machine learning with statistical imputation for predicting drug approvals. Harv. Data Sci. Rev. 2019, 1, 1. [Google Scholar] [CrossRef]
- Brownlee, J. How to Develop Multivariate Multi-Step Time Series Forecasting Models for Air Pollution. Machine Learning Mastery. Available online: https://machinelearningmastery.com/how-to-develop-machine-learning-models-for-multivariate-multi-step-air-pollution-time-series-forecasting/ (accessed on 24 July 2020).
- Park, Y.; Cho, K.H.; Park, J.; Cha, S.M.; Kim, J.H. Development of early-warning protocol for predicting chlorophyll-a concentration using machine learning models in freshwater and estuarine reservoirs, Korea. Sci. Total Environ. 2015, 502, 31–41. [Google Scholar] [CrossRef]
- Box, G.; Jenkins, G. Time Series Analysis Forecasting and Control, rev. ed.; Holden-Day: Oakland, CA, USA, 1976; pp. 303–305. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2826–2830. [Google Scholar] [CrossRef]
- Buitinck, L.; Louppe, G.; Blondel, M.; Pedregosa, F.; Mueller, A.; Grisel, O.; Niculae, V.; Prettenhofer, P.; Gramfort, A.; Grobler, J.; et al. API design for machine learning software: Experiences from the scikit-learn project. In Proceedings of the ECML PKDD Workshop: Languages for Data Mining and Machine Learning, Prage, Czech Republic, 23–27 September 2013; pp. 108–122. [Google Scholar]
- McKinney, W.; Perktold, J.; Seabold, S. Time Series Analysis in Python with Statsmodels. In Proceedings of the 10th Python in Science Conference, Austin, TX, USA, 11−16 July 2011; Walt, S., van der Millman, J., Eds.; Scipy: Austin, TX, USA, 2011; pp. 107–113. [Google Scholar] [CrossRef] [Green Version]
- Lemenkova, P. Processing Oceanographic Data by Python Libraries Numpy, Scipy and Pandas. Aquat. Res. 2019, 2, 73–91. [Google Scholar] [CrossRef]
- Wiltshire, K.H.; Dürselen, C.-D. Revision and Quality Analyses of the Helgoland Reede Long-Term Phytoplankton Data Archive. Helgol. Mar. Res. 2004, 58, 252–268. [Google Scholar] [CrossRef] [Green Version]
- Beutler, M.; Wiltshire, K.H.; Meyer, B.; Moldaenke, C.; Lüring, C.; Meyerhöfer, M.; Hansen, U.-P.; Dau, H. A fluorometric method for the differentiation of algal populations in vivo and in situ. Photosynth. Res. 2002, 72, 39–53. [Google Scholar] [CrossRef]
- Wiltshire, K.H.; Malzahn, A.M.; Wirtz, K.; Greve, W.; Janisch, S.; Mangelsdorf, P.; Manly, B.F.J.; Boersma, M. Resilience of North Sea Phytoplankton Spring Bloom Dynamics: An Analysis of Long-Term Data at Helgoland Roads. Limnol. Oceanogr. 2008, 53, 1294–1302. [Google Scholar] [CrossRef] [Green Version]
- Raabe, T.; Wiltshire, K.H. Quality Control and Analyses of the Long-Term Nutrient Data from Helgoland Roads, North Sea. J. Sea Res. 2009, 61, 3–16. [Google Scholar] [CrossRef]
- Deutsche Wetterdienst (DWD) Climate Data Center (CDC). Daily Station Observations of Sunshine Duration in Hours for Germany. 2020. Available online: https://cdc.dwd.de/portal/ (accessed on 26 April 2020).
- Deutsche Wetterdienst (DWD) Climate Data Center (CDC). Hourly Mean of Station Observations of Wind Speed ca. 10 m above Ground in m/s for Germany. Available online: https://cdc.dwd.de/portal/ (accessed on 6 February 2020).
- Deutsche Wetterdienst (DWD) Climate Data Center (CDC). Hourly Station Observations of Wind Direction 10 m above Ground in Degree for Germany. Available online: https://cdc.dwd.de/portal/ (accessed on 6 February 2020).
- Greve, W.; Reiners, F.; Nast, J.; Hoffmann, S. Helgoland Roads Meso- and Macrozooplankton Time-Series 1974 to 2004: Lessons from 30 Years of Single Spot, High Frequency Sampling at the Only off-Shore Island of the North Sea. Helgol. Mar. Res. 2004, 58, 274–288. [Google Scholar] [CrossRef] [Green Version]
- Irwin, A.J.; Finkel, Z.V. Mining a Sea of Data: Deducing the Environmental Controls of Ocean Chlorophyll. PLoS ONE 2008, 3, e3836. [Google Scholar] [CrossRef] [PubMed]
- Capuzzo, E.; Lynam, C.P.; Barry, J.; Stephens, D.; Forster, R.M.; Greenwood, N.; McQuatters-Gollop, A.; Silva, T.; van Leeuwen, S.M.; Engelhard, G.H. A Decline in Primary Production in the North Sea over 25 Years, Associated with Reductions in Zooplankton Abundance and Fish Stock Recruitment. Glob. Chang. Biol. 2018, 24, e352–e364. [Google Scholar] [CrossRef] [PubMed]
- Scharfe, M.; Wiltshire, K.H. Modeling of Intra-Annual Abundance Distributions: Constancy and Variation in the Phenology of Marine Phytoplankton Species over Five Decades at Helgoland Roads (North Sea). Ecol. Model. 2019, 404, 46–60. [Google Scholar] [CrossRef]
- Mao, H.; Meng, J.; Ji, F.; Zhang, Q.; Fang, H. Comparison of Machine Learning Regression Algorithms for Cotton Leaf Area Index Retrieval Using Sentinel-2 Spectral Bands. Appl. Sci. 2019, 9, 1459. [Google Scholar] [CrossRef] [Green Version]
- Tsai, C.-F.; Hsiao, Y.-C. Combining Multiple Feature Selection Methods for Stock Prediction: Union, Intersection, and Multi-Intersection Approaches. Decis. Support. Syst. 2010, 50, 258–269. [Google Scholar] [CrossRef]
- Lee, J.; Wang, W.; Harrou, F.; Sun, Y. Wind Power Prediction Using Ensemble Learning-Based Models. IEEE Access 2020, 8, 61517–61527. [Google Scholar] [CrossRef]
- Saberioon, M.; Brom, J.; Nedbal, V.; Souček, P.; Císař, P. Chlorophyll-a and Total Suspended Solids Retrieval and Mapping Using Sentinel-2A and Machine Learning for Inland Waters. Ecol. Indic. 2020, 113, 106236. [Google Scholar] [CrossRef]
- Tang, W.; Li, Z.; Cassar, N. Machine Learning Estimates of Global Marine Nitrogen Fixation. J. Geophys. Res. Biogeosci. 2019, 124, 717–730. [Google Scholar] [CrossRef]
- Lenert, M.C.; Walsh, C.G. Balancing Performance and Interpretability: Selecting Features with Bootstrapped Ridge Regression. AMIA Annu. Symp. Proc. 2018, 2018, 1377–1386. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Cortes, C.; Vapnik, V. Support-Vector Networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Hornik, K.; Stinchcombe, M.; White, H. Multilayer Feedforward Networks Are Universal Approximators. Neural Netw. 1989, 2, 359–366. [Google Scholar] [CrossRef]
- Gardner, M.W.; Dorling, S.R. Artificial Neural Networks (the Multilayer Perceptron)—A Review of Applications in the Atmospheric Sciences. Atmos. Environ. 1998, 32, 2627–2636. [Google Scholar] [CrossRef]
- Phung, V.H.; Rhee, E.J. A High-Accuracy Model Average Ensemble of Convolutional Neural Networks for Classification of Cloud Image Patches on Small Datasets. Appl. Sci. 2019, 9, 4500. [Google Scholar] [CrossRef] [Green Version]
- Ooi, K.S.; Chen, Z.; Poh, P.E.; Cui, J. BOD5 Prediction Using Machine Learning Methods. Water Supply 2021, ws2021202. [Google Scholar] [CrossRef]
- Sun, Y.; Li, J.; Liu, J.; Chow, C.; Sun, B.; Wang, R. Using Causal Discovery for Feature Selection in Multivariate Numerical Time Series. Mach. Learn. 2015, 101, 377–395. [Google Scholar] [CrossRef]
- Lee, S.; Chung, J.Y. The Machine Learning-Based Dropout Early Warning System for Improving the Performance of Dropout Prediction. Appl. Sci. 2019, 9, 3093. [Google Scholar] [CrossRef] [Green Version]
- Rezaie-Balf, M.; Kisi, O.; Chua, L.H.C. Application of Ensemble Empirical Mode Decomposition Based on Machine Learning Methodologies in Forecasting Monthly Pan Evaporation. Hydrol. Res. 2019, 50, 498–516. [Google Scholar] [CrossRef]
- Schloen, J.; Stanev, E.V.; Grashorn, S. Wave-Current Interactions in the Southern North Sea: The Impact on Salinity. Ocean. Model. 2017, 111, 19–37. [Google Scholar] [CrossRef]
- Chen, W.-H.; Hsu, S.-H.; Shen, H.-P. Application of SVM and ANN for Intrusion Detection. Comput. Oper. Res. 2005, 32, 2617–2634. [Google Scholar] [CrossRef]
- Munson, M.A.; Caruana, R. On Feature Selection, Bias-Variance, and Bagging. In Machine Learning and Knowledge Discovery in Databases; Buntine, W., Grobelnik, M., Mladenić, D., Shawe-Taylor, J., Eds.; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2009; Volume 5782, pp. 144–159. ISBN 978-3-642-04173-0. [Google Scholar]
- Blauw, A.N.; Benincà, E.; Laane, R.W.P.M.; Greenwood, N.; Huisman, J. Predictability and Environmental Drivers of Chlorophyll Fluctuations Vary across Different Time Scales and Regions of the North Sea. Progress Oceanogr. 2018, 161, 1–18. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).