Abstract
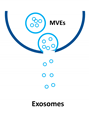
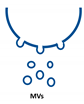
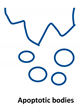
Extracellular vesicles (EVs) are important players in the communication between different kinds of cells by delivering their content, consisting of different types of RNA, proteins, bioactive lipids, or signaling nucleotides, into their target cells. Several types of EVs are distinguished: (1) exosomes with sizes from 30 to 150 nm originate from the endosomal pathway and form intracellular multivesicular bodies (MVBs), which fuse to the plasma membrane before their secretion. (2) EVs with sizes ranging from 100 to 1000 nm in diameter are formed during cell surface budding. (3) Apoptotic bodies with diameters from 500 to 2000 nm are released from blebbing of the cell membrane of apoptotic cells. It is well established that various RNA molecules such as coding RNAs and noncoding RNAs (long noncoding RNAs, microRNAs, circular RNAs, and rRNAs) are present in different amounts in EVs depending on the type and origin of EV. Here we will give an overview of methods to isolate different types of EVs and to quantify and characterize different RNA species.
1. Introduction
Extracellular vesicles (EVs) are a heterogenous family of membrane-limited vesicles, which are distinguished by their different origins and sizes. EVs released from viable cells are microvesicles (MVs or ectosomes), formed by plasma membrane budding with sizes from 100 to 1000 nm and exosomes originating from the endosomal compartment by fusion of multivesicular bodies with the plasma membrane with a size range from 30 to 150 nm. Apoptotic bodies and vesicles are produced during apoptotic cell death and have diameters from 500 to 2000 nm [1,2]. However, there is some controversy regarding the nomenclature and sizes of different types of vesicles, and one should be careful in using size alone in defining different types of vesicles [3,4]. In the future, other criteria such as mode of biogenesis or the method of isolation will be important features to characterize different kinds of vesicles [5].
The release of EVs was first described as a disposal mechanism to discard unwanted materials from cells. Later studies have shown that EVs are surrounded by a lipid bilayer and shuttle various biological components such as proteins, different types of nucleic acid, and lipids between secreting and recipient cells to promote angiogenic, proinflammatory, and immune responses in target cells [6,7,8,9,10].
The content of EVs and their biological function depends on the cell of origin. Vesicles derived from B-cells and dendritic cells have immune-stimulatory and antitumor effects in vivo, whereas tumor-cell-derived EVs stimulate tumor growth and angiogenesis, and platelet-derived EVs promote tumor progression and metastasis of lung cancer cells [11,12,13]. Vesicles have been isolated from diverse bodily fluids, including blood, urine, ascites fluid, and cerebrospinal fluid [14,15,16,17]. Because EVs get their content from their origin cells and, additionally, the number of EVs increase in different human diseases, EVs in blood are currently discussed as novel biomarkers. For example, levels of EVs containing tumor-associated proteins increase in blood samples of cancer patients and correlate with the clinical outcome [18,19]. Moreover, EVs are not only important for diagnostic but also for therapeutic purposes by using exosome or exosome mimetics, such as liposomes, for targeted drug delivery by loading a drug or a detectable agent into appropriate vesicles [20,21,22]. Circulating vesicles are composed of different types of EVs. Accordingly, it was demonstrated that a single cell type can release both exosomes and MVs [23,24].
However, currently available purification methods do not allow fully discriminating between these different types of EVs, and a major ongoing challenge is to establish methods that will allow clearly distinguishing between exosomes and MVs.
Extracellular RNA (exRNA) has been identified as a prothrombotic and proinflammatory factor, which is released from cells under pathological conditions by active and passive processes [25,26,27,28,29,30,31,32]. exRNA is protected from ribonucleolytic digestion within EVs and other vehicles such as lipoproteins, ribonucleoprotein particles, or protein complexes [8,33,34,35]. In this review, we will focus on the isolation and characterization of EVs and types of RNA identified in different types of EVs.
2. Isolation and Characterization of EVs
Methods employed so far for the isolation of EVs from blood or cell culture supernatants are mostly based on several centrifugation steps, size exclusion chromatography, filtration, precipitation, or the use of magnetic or agarose beads. In most studies, EVs are isolated from the supernatants of cultured cells or from blood samples by performing differential centrifugation steps, whereby at centrifugal forces of 200–1500× g, cells and cellular debris of 10,000–20,000× g vesicles with a size between 100 and 800 nm (microvesicles) and between 100,000× and 200,000× g, vesicles with a diameter <100 nm (exosomes) were pelleted [36]. Additionally, filtration has been used to remove larger vesicles from smaller ones [36]. Furthermore, the presence of characteristic surface proteins on certain EVs is eligible for immunoaffinity isolation or magnetic activating cell sorting (MACS) [37,38,39]. For the isolation of exosomes, the highest yields were produced by using the method of density gradient ultracentrifugation in comparison to the method using differential ultracentrifugation [40]. However, stepped ultracentrifugation procedures cannot achieve absolute separation by size because sedimentation also depends on other parameters such as the type of anticoagulant used, the centrifugal speed to yield platelet-free plasma, the sample viscosity, or the type of rotor used [4,41]. A variety of further methods to enrich EVs has been described by groups of the Extracellular RNA Communication Consortium, which include methods of density gradients, precipitation, and filtration such as membrane and gel-filtration-based methods. Using syringe filters with successively smaller pore sizes, EVs of different sizes from cell culture supernatants or biofluids were separated, whereas the method of gel filtration or size exclusion chromatography is based on columns containing a solid phase consisting of porous beads that retains smaller molecules whereby larger molecules cannot enter the pores and are eluted from the column quickly [42]. The method of affinity purification, whereby the solid phase is coupled to specific antigens or other molecules present on the surface of EVs, is mostly used to isolate EVs that express disease-specific antigens [4,42,43]. Furthermore, a number of commercially available kits were developed to isolate exosomes and MVs, but these kits often fail to distinguish between EVs and other membrane-free macromolecular particles such as protein aggregates [1].
Isolated MVs were characterized by several methods that were reviewed by Witwer et al. [4]. Mostly, EVs isolated by differential centrifugation were subsequently studied by flow cytometry [44], whereby scattering flow cytometry requires bead calibration with polystyrene/latex microspheres of known size. However, the detection limit of this method is greater than or equal to 300 nm [45]. Fluorescence-activated cell sorting (FACS) allows classification and specific sorting based on the fluorescence signal of EVs. Although special methods of flow cytometry have been described, the limitation of flow cytometry is its ability to sort EVs <130 nm [5]. The gold-standard method to determine the size of vesicles remains transmission electron microscopy (TEM), which has proven very useful in EV research, as it detects not only the size but also the morphology of isolated vesicles [41]. Furthermore, membrane-surrounded EVs can be recognized and distinguished from other particles such as ribonucleoprotein particles (RNPs) or lipoproteins [1,4]. In general, concentrated EV suspensions are applied to fixed grids, and membrane-surrounded vesicles are recognized by staining with heavy-metal stains such as osmium tetroxide or uranyl acetate [4]. TEM is frequently combined with the use of specific antibodies coupled to nanogold particles for further specification of isolated EVs. Atomic force microscopy is used to further study EV morphology, whereby a mechanical cantilever is passed over a surface, and the presence of surface structures, such as surface-bound particles, can be detected [46]. Furthermore, nanoparticle tracking analysis is useful to determine the concentration and size of vesicles by using a laser beam, which is scattered by particles in the sample [47]. Further characterization of EVs relies on different protein components of different types of EVs and includes biochemical methods such as immunoblotting, Western blot analysis, proteomic analysis, or imaging techniques. For example, exosomes contain endosome-associated proteins, including tetraspanins such as CD9, CD63, CD81, or CD82, which have been used to identify these vesicles [48,49]. However, new data indicate that some proteins regarded as exosome markers (e.g., heat-shock proteins, flotillins, and major histocompatibility complex) are present in all different EV types, and additionally, a number of subpopulations, especially of exosomes, with diverse morphology and distinct biological functions, have been described [50,51,52,53,54].
In summary, most of the strategies used for the isolation of EVs are still under development. Fractions obtained after differential centrifugation steps always contain proteins characteristic of other fractions [43]. Furthermore, EVs are rapidly cleared from peripheral blood, and thus strategies need to be developed to stabilize them [55]. The number of EVs isolated from plasma is also dependent on the type of anticoagulant used for the generation of plasma samples [4,42]. Lipoproteins, microbes, microsomes, or protein aggregates may be copurified in vesicle preparations.
3. Characterization of RNA in Extracellular Vesicles
EVs contain various biotypes of RNA that represent a selected portion of the RNA content of the source cell, including noncoding RNA (ncRNA) such as microRNA (miRNA), fragmented and intact mRNA, ribosomal RNA (rRNA), and long noncoding RNA (lncRNA).
Several reports have demonstrated that genetic exchange of miRNA or mRNA between cells can be accomplished through microvesicle- or exosome-mediated transfer [34,56]. It was reported that circulating EVs in the plasma are mostly derived from platelets and contain mainly miRNAs, whereby many of these miRNAs are predicted to regulate hematopoiesis and cellular differentiation [57]. Furthermore, endothelial progenitor cell-derived microvesicles activate an angiogenic program in endothelial cells by the horizontal transfer of mRNA [23]. Tumor-derived MVs transfer mRNA to monocytes to activate these cells to produce cytokines, which are involved to enhance tumor growth and dampen the immune response [58]. Accordingly, glioblastoma tumor cells release exosomes containing mRNA, miRNA, but no rRNA, and angiogenic proteins and are able to deliver genetic information to recipient cells in the tumor environment [8]. Additionally, exosomes and special types and sequences of extracellular MV-associated RNAs were useful biomarkers, for example for monitoring prostate cancer invasion and metastasis [8,59,60].
Cellular mechanisms involved in the package of different types of RNA into different types of EVs and their regulated uptake into specific target molecules are still under investigation [61]. Differences in the EV-RNA versus the intracellular RNA profile were described, and it has been suggested that several EV-sorting mechanisms such as different stabilities of special RNA species, which might result from the presence of RNA-binding proteins or special RNases in EVs, must be involved [8,59,62,63,64,65]. Furthermore, special nuclear ribonucleoproteins were suggested to control the sorting of miRNAs into exosomes through binding to specific motifs [66]. RNA loading in EVs further depends on cell activation or pathological conditions. For instance, it is well established that stressors such as hypoxia alter the release of EVs and the RNA profile in EVs released in comparison to the donor cells [67].
RNA concentrations in biofluids are very low, 20–50 ng/mL of total plasma, and 5 ng/mL of EV-associated RNA, and several methods have been attempted aiming to quantify and identify different kinds of extracellular RNA in EVs [42,68].
As expected, the quantity and quality of EV-RNA depend on the method used for EV isolation [40,43,69,70]. Furthermore, the method used for RNA isolation also has an influence on the yield and quality of RNA [42]. In most studies, quantification of isolated RNA is performed using NanoDrop, the Qubit RNA assay, or a bioanalyzer (Agilent), which differ in their detection limit [43]. Only the bioanalyzer, which has the lowest range limit of 50 pg/mL, is able to evaluate size distributions of the RNA molecules additionally. Quantitative real-time polymerase chain reaction (qRT-PCR) is another valuable quantification method [42]. Further molecular analysis of exRNA includes methods such as NanoString, microarray, and next-generation sequencing (NGS) [42]. For further quantitative profiling of small RNAs such as miRNAs, piwi-interacting RNAs (piRNAs), or small nucleolar (sno) RNAs, small RNA sequencing using RNA spike-in molecules is used [71,72].
So far, most current studies have used microarray analysis and qRT-PCR to examine EV-RNA with a focus on miRNAs. For example, exosomes have been found to contain mainly functional mRNAs and miRNAs [34]. Because unknown miRNAs or other RNA species are often undetectable, a sequencing-based RNA profiling analysis from exosomes isolated from blood samples demonstrated that plasma-derived exosomes not only contain miRNA and mRNA but also a large variety of other small noncoding RNA species, including RNA transcript, repeat sequences, structural RNAs, and tRNA fragments [73,74,75]. Furthermore, deep sequencing or RNA in exosomes from the colon cancer cell line identified special miRNA sequences enriched in EVs compared to the parent cancer cell [76]. In future, the use of deep sequencing will ensure the characterization of the whole transcriptome of blood-derived exosomes. However, the distribution of different kinds of RNA in EVs is highly dependent on the method or kit used for the isolation of EVs, and the co-precipitation of non-EV microparticles or RNA-binding proteins should be excluded [74].
Size filtration of conditioned medium from glioblastoma stem-like cell cultures reveals that mRNA is enriched in MVs, whereas exosomes have the highest fraction of miRNA [42,72]. Using the method of differential centrifugation, the RNA profile showed that rRNA was preferred detectable in apoptotic bodies and smaller RNAs, but no rRNAs were present in exosomes. In these studies, detection, quality, yield, and size of vesicular RNAs were analyzed using capillary electrophoresis by the bioanalyzer (Agilent) [48]. Accordingly, rRNA and additionally DNA are mainly present in microparticles from cells undergoing apoptosis in vitro [77,78]. Table 1 summarizes types of RNA isolated from different EV populations.

Table 1.
Type of RNA in different EVs. Exosomes (30–150 nm) originate from the endosomal pathway by fusion of multivesicular bodies (MVBs) with the plasma membrane, microvesicles (MVs, 100–1000 nm) are formed by cell surface membrane blebbing, and apoptotic bodies (500–2000 nm) are vesicles released from apoptotic cells.
Type of RNA in EVs is furthermore dependent on the source used for EV isolation. Subpopulations of EVs isolated from metastatic tumor tissue showed distinct size and morphology and differed in their RNA cargo [79]. Type of RNA identified in different EVs isolated from cell culture supernatants was not only dependent on the type of donor cell but also on the type of stimulation [48,80]. After culturing the mast cell line HMC-1 in normal growth medium, MVs isolated from supernatants did not contain rRNA, but when HMC-1 was treated with degranulating agents such as ionomycin or complement factor in serum-free medium, MVs contained high amounts of rRNA and fewer small RNAs [48,80]. Other studies confirmed that the type of RNA, present in EVs isolated from cell-culture-conditioned medium, is dependent on culture conditions. For example, bovine serum also contains RNA in EVs, and methods to remove exRNA from bovine serum through ultracentrifugation or affinity purification are only partly successful so far [81,82].
In summary, many studies failed to demonstrate whether identified extracellular RNAs were associated with EVs or rather with RNA–protein complexes, viral particles, RNPs, and lipoproteins (HDL and LDL) that might have been co-isolated with EVs [1,43]. However, in this regard, it has been reported that EVs isolated by ultracentrifugation may become aggregated and are contaminated by nonvesicular macromolecules. Additional purification techniques such as high-resolution density gradient fractionation and direct immunoaffinity capture techniques were used to characterize EVs and nonvesicular material precisely [83]. DNA known to be present in several biological fluids was also demonstrated to be present in EV-RNA preparations and thus can interfere with the downstream analysis of RNA [84]. rRNA might also become associated with EVs during centrifugation steps. This might explain the variable presence of rRNA in EVs described in the literature, and the question of whether rRNAs are naturally associated with EVs or if they are impurities or contaminants is still to be discussed [43]. To remove outside bound DNA and RNA and destroy protein complexes such as RNPs, EVs that will be used for the isolation of RNA have to be pretreated with DNase, RNase, and proteinase [43]. Small RNA-seq analysis has indicated that EVs are associated with various fragments from coding and noncoding RNA, including rRNA, tRNA, snRNA, lncRNA, and vault RNA [65,75,85]. Currently, there is no knowledge whether these fragments are artifacts produced during the isolation or are formed by specific processing steps either in the cytoplasm of donor cells or inside the vesicles [86].
4. Conclusions
EVs are identified as a very important tool in almost all biological processes in the body by facilitating the cross-talk between different cells, resulting in maintaining tissue homeostasis and angiogenesis and in pathologies such as cancerogenesis, chronic inflammation, or atherosclerosis. So far, different methods have been used to isolate EVs and to characterize RNA inside these vesicles. The development of standardized approaches is needed to obtain reproducible and comparable results that will allow the implementation of EVs as clinical biomarkers or their usefulness as drug delivery devices.
Author Contributions
Writing—original draft preparation, S.F.; review and editing, S.F. and E.D.; Both authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [Green Version]
- Vidal, M. Exosomes: Revisiting their role as “garbage bags”. Traffic 2019, 20, 815–828. [Google Scholar] [CrossRef] [PubMed]
- Gould, S.J.; Raposo, G. As we wait: Coping with an imperfect nomenclature for extracellular vesicles. J. Extracell. Vesicles 2013, 2, 20389. [Google Scholar] [CrossRef] [PubMed]
- Witwer, K.W.; Buzás, E.I.; Bemis, L.; Bora, A.; Lässer, C.; Lötvall, J.; Hoen, E.N.N.; Piper, M.G.; Sivaraman, S.; Skog, J.; et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracell. Vesicles 2013, 2, 20360. [Google Scholar] [CrossRef] [PubMed]
- Momen-Heravi, F.; Balaj, L.; Alian, S.; Tigges, J.; Toxavidis, V.; Ericsson, M.; Distel, R.J.; Ivanov, A.R.; Skog, J.; Kuo, W.P. Alternative methods for characterization of extracellular vesicles. Front. Physiol. 2012, 3, 354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maacha, S.; Bhat, A.A.; Jimenez, L.; Raza, A.; Haris, M.; Uddin, S.; Grivel, J.C. Extracellular vesicles-mediated intercellular communication: Roles in the tumor microenvironment and anti-cancer drug resistance. Mol. Cancer 2019, 18, 55. [Google Scholar] [CrossRef] [Green Version]
- Simpson, R.J.; Lim, J.W.; Moritz, R.L.; Mathivanan, S. Exosomes: Proteomic insights and diagnostic potential. Expert Rev. Proteom. 2009, 6, 267–283. [Google Scholar] [CrossRef]
- Skog, J.; Wurdinger, T.; van Rijn, S.; Meijer, D.; Gainche, L.; Sena-Esteves, M.; Curry, W.T.; Carter, R.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and protein that promote tumor growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef]
- Vajen, T.; Mause, S.F.; Koenen, R.R. Microvesicles from platelets: Novel drivers of vascular inflammation. Thromb. Haemost. 2015, 114, 228–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Vos, K.E.; Abels, E.R.; Zhang, X.; Lai, C.; Carrizosa, E.; Oakley, D.; Prabhakar, S.; Mardini, O.; Crommentuijn, M.H.W.; Skog, J.; et al. Directly visualized glioblastoma-derived extracellular vesicles transfer RNA to microglia/macrophages in the brain. Neuro-Oncology 2015, 18, 58–69. [Google Scholar] [CrossRef] [Green Version]
- Chaput, N.; Taïeb, J.; Andre, F.; Zitvogel, L. The potential of exosomes in immunotherapy. Expert Opin. Biol. Ther. 2005, 5, 737–747. [Google Scholar] [CrossRef] [PubMed]
- Janowska-Wieczorek, A.; Wysoczynski, M.; Kijowski, J.; Marquez-Curtis, L.; Machalinski, B.; Ratajczak, J.; Ratajczak, M.Z. Microvesicles derived from activated platelets induce metastasis and angiogenesis in lung cancer. Int. J. Cancer 2004, 113, 752–760. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Yu, S.; Zinn, K.; Wang, J.; Zhang, L.; Jia, Y.; Kappes, J.C.; Barnes, S.; Kimberly, R.; Grizzle, W.E.; et al. Murine Mammary Carcinoma Exosomes Promote Tumor Growth by Suppression of NK Cell Function. J. Immunol. 2006, 176, 1375–1385. [Google Scholar] [CrossRef] [Green Version]
- Andre, F.; Schartz, N.E.; Movassagh, M.; Flament, C.; Pautier, P.; Morice, P.; Pomel, C.; Lhomme, C.; Escudier, B.; Le Chevalier, T.; et al. Malignant effusions and immunogenic tumour-derived exosomes. Lancet 2002, 360, 295–305. [Google Scholar] [CrossRef]
- Caby, M.-P.; Lankar, D.; Vincendeau-Scherrer, C.; Raposo, G.; Bonnerot, C. Exosomal-like vesicles are present in human blood plasma. Int. Immunol. 2005, 17, 879–887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pisitkun, T.; Shen, R.-F.; Knepper, M.A. Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. USA 2004, 101, 13368–13373. [Google Scholar] [CrossRef] [Green Version]
- Vella, L.; Sharples, R.; Lawson, V.; Masters, C.; Cappai, R.; Hill, A. Packaging of prions into exosomes is associated with a novel pathway of PrP processing. J. Pathol. 2007, 211, 582–590. [Google Scholar] [CrossRef]
- Galindo-Hernandez, O.; Villegas-Comonfort, S.; Candanedo, F.; Vázquez, M.C.G.; Chavez-Ocaña, S.; Jimenez-Villanueva, X.; Sierra-Martinez, M.; Salazar, E.P. Elevated Concentration of Microvesicles Isolated from Peripheral Blood in Breast Cancer Patients. Arch. Med. Res. 2013, 44, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Menck, K.; Scharf, C.; Bleckmann, A.; Dyck, L.; Rost, U.; Wenzel, D.; Dhople, V.M.; Siam, L.; Pukrop, T.; Binder, C.; et al. Tumor-derived microvesicles mediate human breast cancer invasion through differentially glycosylated EMMPRIN. J. Mol. Cell Biol. 2014, 7, 143–153. [Google Scholar] [CrossRef] [Green Version]
- Ailuno, G.; Baldassari, S.; Lai, F.; Florio, T.; Caviglioli, G. Exosomes and Extracellular Vesicles as Emerging Theranostic Platforms in Cancer Research. Cells 2020, 9, 2569. [Google Scholar] [CrossRef] [PubMed]
- Antimisiaris, S.G.; Mourtas, S.; Marazioti, A. Exosomes and Exosome-Inspired Vesicles for Targeted Drug Delivery. Pharmacy 2018, 10, 218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnsen, K.B.; Gudbergsson, J.M.; Skov, M.N.; Pilgaard, L.; Moos, T.; Duroux, M. A comprehensive overview of exosomes as drug delivery vehicles—Endogenous nanocarriers for targeted cancer therapy. Biochim. Biophys. Acta 2014, 1846, 75–87. [Google Scholar] [CrossRef]
- Deregibus, M.C.; Cantaluppi, V.; Calogero, R.; Iacono, M.L.; Tetta, C.; Biancone, L.; Bruno, S.; Bussolati, B.; Camussi, G. Endothelial progenitor cell derived microvesicles activate an angiogenic program in endothelial cells by a horizontal transfer of mRNA. Blood 2007, 110, 2440–2448. [Google Scholar] [CrossRef] [Green Version]
- Heijnen, H.F.; Schiel, A.E.; Fijnheer, R.; Geuze, H.J.; Sixma, J.J. Activated platelets release two types of membrane vesicles: Microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha-granules. Blood 1999, 94, 3791–3799. [Google Scholar] [CrossRef] [PubMed]
- Fischer, S.; Gerriets, T.; Wessels, C.; Walberer, M.; Kostin, S.; Stolz, E.; Zheleva, K.; Hocke, A.; Hippenstiel, S.; Preissner, K.T. Extracellular RNA mediates endothelial-cell permeability via vascular endothelial growth factor. Blood 2007, 110, 2457–2465. [Google Scholar] [CrossRef] [Green Version]
- Fischer, S.; Gesierich, S.; Griemert, B.; Schänzer, A.; Acker, T.; Augustin, H.G.; Olsson, A.-K.; Preissner, K.T. Extracellular RNA liberates Tumor-Necrosis-Factor-α to promote tumor cell trafficking and progression. Cancer Res. 2013, 73, 5080–5089. [Google Scholar] [CrossRef] [Green Version]
- Fischer, S.; Grantzow, T.; Pagel, J.-I.; Tschernatsch, M.; Sperandio, M.; Preissner, K.T.; Deindl, E. Extracellular RNA promotes leukocyte recruitment in the vascular system by mobilizing proinflammatory cytokines. Thromb. Haemost. 2012, 108, 730–741. [Google Scholar] [PubMed]
- Fischer, S.; Preissner, K.T. Extracellular nucleic acids as novel alarm signals in the vascular system: Mediators of defence and disease. Hämostaseologie 2013, 33, 37–42. [Google Scholar] [CrossRef]
- Kannemeier, C.; Shibamiya, A.; Nakazawa, F.; Trusheim, H.; Ruppert, C.; Markart, P.; Song, Y.; Tzima, E.; Kennerknecht, E.; Niepmann, M.; et al. Extracellular RNA constitutes a natural procoagulant cofactor in blood coagulation. Proc. Natl. Acad. Sci. USA 2007, 104, 6388–6393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tielking, K.; Fischer, S.; Preissner, K.T.; Vajkoczy, P.; Xu, R. Extracellular RNA in Central Nervous System Pathologies. Front. Mol. Neurosci. 2019, 12, 254. [Google Scholar] [CrossRef] [Green Version]
- Tosar, J.P.; Witwer, K.; Cayota, A. Revisiting Extracellular RNA Release, Processing, and Function. Trends Biochem. Sci. 2021, 46, 438–445. [Google Scholar] [CrossRef]
- Yuan, T.; Huang, X.; Woodcock, M.; Du, M.; Dittmar, R.; Wang, Y.; Tsai, S.; Kohli, M.; Boardman, L.A.; Patel, T.; et al. Plasma extracellular RNA profiles in healthy and cancer patients. Sci. Rep. 2016, 6, 19413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arroyo, J.; Chevillet, J.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.; Bennett, C.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef] [Green Version]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011, 13, 423–433. [Google Scholar] [CrossRef] [Green Version]
- Van der Pol, E.; Böing, A.N.; Harrison, P.; Sturk, A.; Nieuwland, R. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol. Rev. 2012, 64, 676–705. [Google Scholar] [CrossRef] [Green Version]
- Clayton, A.; Court, J.; Navabi, H.; Adams, M.; Mason, M.D.; Hobot, J.A.; Newman, G.R.; Jasani, B. Analysis of antigen presenting cell derived exosomes, based on immuno-magnetic isolation and flow cytometry. J. Immunol. Methods 2001, 247, 163–174. [Google Scholar] [CrossRef]
- Taylor, D.D.; Gercel-Taylor, C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008, 110, 13–21. [Google Scholar] [CrossRef]
- Wubbolts, R.; Leckie, R.S.; Veenhuizen, P.T.; Schwarzmann, G.; Möbius, W.; Hoernschemeyer, J.; Slot, J.W.; Geuze, H.J.; Stoorvogel, W. Proteomic and biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation. J. Biol. Chem. 2003, 278, 10963–10972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Deun, J.; Mestdagh, P.; Sormunen, R.; Cocquyt, V.; Vermaelen, K.; Vandesompele, J.; Bracke, M.; De Wever, O.; Hendrix, A. The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. J. Extracell. Vesicles 2014, 3, 24858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- György, B.; Szabó, T.G.; Pásztói, M.; Pál, Z.; Misják, P.; Aradi, B.; László, V.; Pállinger, E.; Pap, E.; Kittel, A.; et al. Membrane vesicles, current state-of-the-art: Emerging role of extracellular vesicles. Cell. Mol. Life Sci. 2011, 68, 2667–2688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laurent, L.; Abdel-Mageed, A.B.; Adelson, P.D.; Arango, J.; Balaj, L.; Breakefield, X.; Carlson, E.; Carter, B.S.; Majem, B.; Chen, C.C.; et al. Meeting report: Discussions and preliminary findings on extracellular RNA measurement methods from laboratories in the NIH Extracellular RNA Communication Consortium. J. Extracell. Vesicles 2015, 4, 26533. [Google Scholar] [CrossRef] [PubMed]
- Mateescu, B.; Kowal, E.J.K.; Van Balkom, B.W.M.; Bartel, S.; Bhattacharyya, S.N.; Buzás, E.I.; Buck, A.; De Candia, P.; Chow, W.-N.; Das, S.; et al. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA—An ISEV position paper. J. Extracell. Vesicles 2017, 6, 1286095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menck, K.; Bleckmann, A.; Schulz, M.; Ries, L.; Binder, C. Isolation and Characterization of Microvesicles from Peripheral Blood. J. Vis. Exp. 2017, 119, e55057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hein, B.; Willig, K.; Hell, S.W. Stimulated emission depletion (STED) nanoscopy of a fluorescent protein-labeled organelle inside a living cell. Proc. Natl. Acad. Sci. USA 2008, 105, 14271–14276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuana, Y.; Oosterkamp, T.H.; Bahatyrova, S.; Ashcroft, B.; Rodriguez, P.G.; Bertina, R.M.; Osanto, S. Atomic force microscopy: A novel approach to the detection of nanosized blood microparticles. J. Thromb. Haemost. 2010, 8, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Dragovic, R.A.; Gardiner, C.; Brooks, A.S.; Tannetta, D.S.; Ferguson, D.; Hole, P.; Carr, B.; Redman, C.W.; Harris, A.; Dobson, P.J.; et al. Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomed. Nanotechnol. Biol. Med. 2011, 7, 780–788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crescitelli, R.; Lässer, C.; Szabó, T.G.; Kittel, A.; Eldh, M.; Dianzani, I.; Buzás, E.I.; Lötvall, J. Distinct RNA profiles in subpopulations of extracellular vesicles: Apoptotic bodies, microvesicles and exosomes. J. Extracell. Vesicles 2013, 2, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Janockova, J.; Slovinska, L.; Harvanova, D.; Spakova, T.; Rosocha, J. New therapeutic approaches of mesenchymal stem cells-derived exosomes. J. Biomed. Sci. 2021, 28, 39. [Google Scholar] [CrossRef]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lässer, C.; Jang, S.C.; Lötvall, J. Subpopulations of extracellular vesicles and their therapeutic potential. Mol. Asp. Med. 2018, 60, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Willms, E.; Johansson, H.J.; Mäger, I.; Lee, Y.; Blomberg, K.E.M.; Sadik, M.; Alaarg, A.; Smith, C.I.E.; Lehtiö, J.; El Andaloussi, S.; et al. Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci. Rep. 2016, 6, 22519. [Google Scholar] [CrossRef]
- Zhang, H.; Freitas, D.; Kim, H.S.; Fabijanic, K.; Li, Z.; Chen, H.; Mark, M.T.; Molina, H.; Martin, A.B.; Bojmar, L.; et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat. Cell Biol. 2018, 20, 332–343. [Google Scholar] [CrossRef] [PubMed]
- Zabeo, D.; Cvjetkovic, A.; Lässer, C.; Schorb, M.; Lötvall, J.; Höög, J.L. Exosomes purified from a single cell type have diverse morphology. J. Extracell. Vesicles 2017, 6, 1329476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ratajczak, M.Z.; Ratajczak, J. Extracellular microvesicles/exosomes: Discovery, disbelief, acceptance, and the future? Leukemia 2020, 34, 3126–3135. [Google Scholar] [CrossRef] [PubMed]
- Ekström, K.; Valadi, H.; Sjöstrand, M.; Malmhäll, C.; Bossios, A.; Eldh, M.; Lötvall, J. Characterization of mRNA and microRNA in human mast cell-derived exosomes and their transfer to other mast cells and blood CD34 progenitor cells. J. Extracell. Vesicles 2012, 1, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Hunter, M.P.; Ismail, N.; Zhang, X.; Aguda, B.D.; Lee, E.J.; Yu, L.; Xiao, T.; Schafer, J.; Lee, M.-L.T.; Schmittgen, T.D.; et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS ONE 2008, 3, e3694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baj-Krzyworzeka, M.; Szatanek, R.; Węglarczyk, K.; Baran, J.; Zembala, M. Tumour-derived microvesicles modulate biological activity of human monocytes. Immunol. Lett. 2007, 113, 76–82. [Google Scholar] [CrossRef]
- Gao, Z.; Pang, B.; Li, J.; Gao, N.; Fan, T.; Li, Y. Emerging Role of Exosomes in Liquid Biopsy for Monitoring Prostate Cancer Invasion and Metastasis. Front. Cell Dev. Biol. 2021, 9, 679527. [Google Scholar] [CrossRef] [PubMed]
- Gui, Y.; Liu, H.; Zhang, L.; Lv, W.; Hu, X. Altered microRNA profiles in cerebrospinal fluid exosome in Parkinson disease and Alzheimer disease. Oncotarget 2015, 6, 37043–37053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606. [Google Scholar] [CrossRef] [PubMed]
- Batagov, A.O.; Kurochkin, I.V. Exosomes secreted by human cells transport largely mRNA fragments that are enriched in the 3′-untranslated regions. Biol. Direct 2013, 8, 12. [Google Scholar] [CrossRef] [Green Version]
- Chai, Y.; Liu, J.; Zhang, Z.; Liu, L. HuR-regulated lncRNA NEAT1 stability in tumorigenesis and progression of ovarian cancer. Cancer Med. 2016, 5, 1588–1598. [Google Scholar] [CrossRef] [PubMed]
- Noerholm, M.; Balaj, L.; Limperg, T.; Salehi, A.; Zhu, L.D.; Hochberg, F.H.; Breakefield, X.O.; Carter, B.S.; Skog, J. RNA expression patterns in serum microvesicles from patients with glioblastoma multiforme and controls. BMC Cancer 2012, 12, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Balkom, B.W.M.; Eisele, A.S.; Pegtel, D.M.; Bervoets, S.; Verhaar, M. Quantitative and qualitative analysis of small RNAs in human endothelial cells and exosomes provides insights into localized RNA processing, degradation and sorting. J. Extracell. Vesicles 2015, 4, 26760. [Google Scholar] [CrossRef] [PubMed]
- Villarroya-Beltri, C.; Gutierrez-Vazquez, C.; Sanchez-Cabo, F.; Pérez-Hernández, D.; Vázquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sánchez-Madrid, F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013, 4, 2980. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.; Zhang, J.-J.; Peng, Y.-P.; Zhu, Y.; Yin, L.-D.; Wei, J.-S.; Gao, W.-T.; Jiang, K.-R.; Miao, Y. A Yin-Yang 1/miR-30a regulatory circuit modulates autophagy in pancreatic cancer cells. J. Transl. Med. 2017, 15, 211. [Google Scholar] [CrossRef] [Green Version]
- Williams, Z.; Ben-Dov, I.; Elias, R.; Mihailovic, A.; Brown, M.; Rosenwaks, Z.; Tuschl, T. Comprehensive profiling of circulating microRNA via small RNA sequencing of cDNA libraries reveals biomarker potential and limitations. Proc. Natl. Acad. Sci. USA 2013, 110, 4255–4260. [Google Scholar] [CrossRef] [Green Version]
- Eldh, M.; Lötvall, J.; Malmhäll, C.; Ekström, K. Importance of RNA isolation methods for analysis of exosomal RNA: Evaluation of different methods. Mol. Immunol. 2012, 50, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Rekker, K.; Saare, M.; Roost, A.M.; Kubo, A.-L.; Zarovni, N.; Chiesi, A.; Salumets, A.; Peters, M. Comparison of serum exosome isolation methods for microRNA profiling. Clin. Biochem. 2014, 47, 135–138. [Google Scholar] [CrossRef]
- Giraldez, M.D.; Spengler, R.M.; Etheridge, A.; Godoy, P.M.; Barczak, A.J.; Srinivasan, S.; De Hoff, P.L.; Tanriverdi, K.; Courtright, A.; Lu, S.; et al. Comprehensive multi-center assessment of small RNA-seq methods for quantitative miRNA profiling. Nat. Biotechnol. 2018, 36, 746–757. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Batagov, A.O.; Schinelli, S.; Wang, J.; Wang, Y.; El Fatimy, R.; Rabinovsky, R.; Balaj, L.; Chen, C.C.; Hochberg, F.; et al. Coding and noncoding landscape of extracellular RNA released by human glioma stem cells. Nat. Commun. 2017, 8, 1145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bellingham, S.A.; Coleman, B.M.; Hill, A.F. Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells. Nucleic Acids Res. 2012, 40, 10937–10949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.; Yuan, T.; Tschannen, M.; Sun, Z.; Jacob, H.; Du, M.; Liang, M.; Dittmar, R.L.; Liu, Y.; Liang, M.; et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genom. 2013, 14, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Hoen, E.N.M.N.; Buermans, H.P.J.; Waasdorp, M.; Stoorvogel, W.; Wauben, M.; Hoen, P. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012, 40, 9272–9285. [Google Scholar] [CrossRef] [Green Version]
- Ji, H.; Chen, M.; Greening, D.W.; He, W.; Rai, A.; Zhang, W.; Simpson, R.J. Deep sequencing of RNA from three different extracellular vesicle (EV) subtypes released from the human LIM1863 colon cancer cell line uncovers distinct miRNA-enrichment signatures. PLoS ONE 2014, 9, e110314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reich, C.F.; Pisetsky, D.S. The content of DNA and RNA in microparticles released by Jurkat and HL-60 cells undergoing in vitro apoptosis. Exp. Cell Res. 2009, 315, 760–768. [Google Scholar] [CrossRef]
- Spencer, D.M.; Gauley, J.; Pisetsky, D.S. The properties of microparticles from RAW 264.7 macrophage cells undergoing in vitro activation or apoptosis. Innate Immun. 2013, 20, 239–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crescitelli, R.; Lässer, C.; Jang, S.C.; Cvjetkovic, A.; Malmhäll, C.; Karimi, N.; Höög, J.L.; Johansson, I.; Fuchs, J.; Thorsell, A.; et al. Subpopulations of extracellular vesicles from human metastatic melanoma tissue identified by quantitative proteomics after optimized isolation. J. Extracell. Vesicles 2020, 9, 1722433. [Google Scholar] [CrossRef]
- Elsemüller, A.K.; Tomalla, V.; Gärtner, U.; Troidl, K.; Jeratsch, S.; Graumann, J.; Baal, N.; Hackstein, H.; Lasch, M.; Deindl, E.; et al. Characterization of mast cell-derived rRNA-containing microvesicles and their inflammatory impact on endothelial cells. FASEB J. 2019, 33, 5457–5467. [Google Scholar] [CrossRef] [PubMed]
- Tosar, J.P.; Cayota, A.; Eitan, E.; Halushka, M.K.; Witwer, K.W. Ribonucleic artefacts: Are some extracellular RNA discoveries driven by cell culture medium components? J. Extracell. Vesicles 2017, 6, 1272832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, Z.; Batagov, A.O.; Carter, D.R.F.; Krichevsky, A.M. Fetal Bovine Serum RNA Interferes with the Cell Culture derived Extracellular RNA. Sci. Rep. 2016, 6, 31175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeppesen, D.; Fenix, A.; Franklin, J.L.; Higginbotham, J.N.; Zhang, Q.; Zimmerman, L.J.; Liebler, D.C.; Ping, J.; Liu, Q.; Evans, R.; et al. Reassessment of Exosome Composition. Cell 2019, 177, 428–445.e18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J.; et al. Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. 2014, 24, 766–769. [Google Scholar] [CrossRef] [Green Version]
- Tosar, J.P.; Gámbaro, F.; Sanguinetti, J.; Bonilla, B.; Witwer, K.W.; Cayota, A. Assessment of small RNA sorting into different extracellular fractions revealed by high-throughput sequencing of breast cell lines. Nucleic Acids Res. 2015, 43, 5601–5616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melo, S.; Sugimoto, H.; O’Connell, J.T.; Kato, N.; Villanueva, A.; Vidal, A.; Qiu, L.; Vitkin, E.; Perelman, L.T.; Melo, C.A.; et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 2014, 26, 707–721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).