In Vitro Simulation of Human Colonic Fermentation: A Practical Approach towards Models’ Design and Analytical Tools
Abstract
:1. Introduction
2. Materials and Methods
3. The Colonic Fermentation Process
4. Available Colonic In Vitro Fermentation Models
4.1. Static Colonic Fermentation Models
4.2. Dynamic Colonic Fermentation Models
5. Addressing Colonic In Vitro Digestion Studies
5.1. Faecal Inoculum Collection and Preparation
5.2. Changes in the Microbiota Populations and Their Metabolic Response
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bhagavan, N.; Ha, C.-E. Gastrointestinal digestion and absorption. In Essentials of Medical Biochemistry; Academic Press: Cambridge, MA, USA, 2015; pp. 137–164. [Google Scholar] [CrossRef]
- Engelking, L.R. Carbohydrate digestion. In Textbook of Veterinary Physiological Chemistry; Academic Press: Cambridge, MA, USA, 2015. [Google Scholar] [CrossRef]
- Engelking, L.R. Lipid digestion. In Textbook of Veterinary Physiological Chemistry; Ademic Press: Cambridge, MA, USA, 2015. [Google Scholar] [CrossRef]
- Bhutia, Y.D.; Ganapathy, V. Protein digestion and absorption. In Physiology of the Gastrointestinal Tract; Ademic Press: Cambridge, MA, USA, 2018; pp. 1063–1086. [Google Scholar]
- Macfarlane, G.T.; Macfarlane, S. Models for intestinal fermentation: Association between food components, delivery systems, bioavailability and functional interactions in the gut. Curr. Opin. Biotechnol. 2007, 18, 156–162. [Google Scholar] [CrossRef]
- Arrieta, M.-C.; Walter, J.; Finlay, B.B. Human Microbiota-Associated Mice: A Model with Challenges. Cell Host Microbe 2016, 19, 575–578. [Google Scholar] [CrossRef] [Green Version]
- Venema, K.; Abbeele, P.V.D. Experimental models of the gut microbiome. Best Pract. Res. Clin. Gastroenterol. 2013, 27, 115–126. [Google Scholar] [CrossRef]
- Calvo-Lerma, J.; Paz-Yépez, C.; Asensio-Grau, A.; Heredia, A.; Andrés, A. Impact of processing and intestinal conditions on in vitro digestion of chia (Salvia hispanica) seeds and derivatives. Foods 2020, 9, 290. [Google Scholar] [CrossRef] [Green Version]
- Asensio-Grau, A.; Calvo-Lerma, J.; Heredia, A.; Andrés, A. In vitro digestion of salmon: Influence of processing and intestinal conditions on macronutrients digestibility. Food Chem. 2021, 342, 128387. [Google Scholar] [CrossRef] [PubMed]
- Asensio-Grau, A.; Calvo-Lerma, J.; Heredia, A.; Andrés, A. Fat digestibility in meat products: Influence of food structure and gastrointestinal conditions. Int. J. Food Sci. Nutr. 2018, 70, 530–539. [Google Scholar] [CrossRef] [PubMed]
- Payne, A.N.; Zihler, A.; Chassard, C.; Lacroix, C. Advances and perspectives in in vitro human gut fermentation modeling. Trends Biotechnol. 2012, 30, 17–25. [Google Scholar] [CrossRef]
- Fehlbaum, S.; Chassard, C.; Schwab, C.; Voolaid, M.; Fourmestraux, C.; Derrien, M.; Lacroix, C. In vitro study of Lac-tobacillus paracasei CNCM I-1518 in healthy and Clostridium difficile colonized elderly gut microbiota. Front. Nutr. 2019, 6, 184. [Google Scholar] [CrossRef] [PubMed]
- Mulet-Cabero, A.I.; Egger, L.; Portmann, R.; Ménard, O.; Marze, S.; Minekus, M.; Golding, M. A standardised semi-dynamic in vitro digestion method suitable for food—An international consensus. Food Funct. 2020, 11, 1702–1720. [Google Scholar] [CrossRef] [Green Version]
- Brodkorb, A.; Egger, L.; Alminger, M.; Alvito, P.; Assunção, R.; Ballance, S.; Clemente, A. INFOGEST static in vitro simulation of gastrointestinal food digestion. Nat. Protoc. 2019, 14, 991–1014. [Google Scholar] [CrossRef]
- Flint, H.J. Who inhabits our gut? In Why Gut Microbes Matter; Springer: Cham, Switzerland, 2020; pp. 47–61. [Google Scholar]
- Wang, T.; Roest, D.I.M.; Smidt, H.; Zoetendal, E.G. “We are what we eat”: How diet impacts the gut microbiota in adulthood. In How Fermented Foods Feed a Healthy Gut Microbiota; Springer: Cham, Switzerland, 2019; pp. 259–283. [Google Scholar]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Louis, P.; Hold, G.L.; Flint, H.J. The gut microbiota, bacterial metabolites, and colorectal cancer. Nat. Rev. Microbiol. 2014, 12, 661–672. [Google Scholar] [CrossRef]
- Scott, K.P.; Gratz, S.W.; Sheridan, P.O.; Flint, H.J.; Duncan, S.H. The influence of diet on the gut microbiota. Pharmacol. Res. 2013, 69, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Chang, H.W.; Yan, D.; Lee, K.M.; Ucmak, D.; Wong, K.; Bhutani, T. Influence of diet on the gut micro-biome and implications for human health. J. Transl. Med. 2017, 15, 73. [Google Scholar] [CrossRef] [Green Version]
- Pandey, K.R.; Naik, S.R.; Vakil, B.V. Probiotics, prebiotics, and synbiotics—A review. J. Food Sci. Technol. 2015, 52, 7577–7587. [Google Scholar] [CrossRef] [PubMed]
- Jamar, G.; Ribeiro, D.A.; Pisani, L.P. High-fat or high-sugar diets as trigger inflammation in the microbio-ta-gut-brain axis. Crit. Rev. Food Sci. Nutr. 2020, 61, 836–854. [Google Scholar] [CrossRef]
- Russell, W.R.; Gratz, S.W.; Duncan, S.H.; Holtrop, G.; Ince, J.; Scobbie, L.; Duthie, G.G. High-protein, re-duced-carbohydrate weight-loss diets promote metabolite profiles likely to be detrimental to colonic health. Am. J. Clin. Nutr. 2011, 93, 1062–1072. [Google Scholar] [CrossRef]
- Machate, D.J.; Figueiredo, P.S.; Marcelino, G.; Guimarães, R.D.C.A.; Hiane, P.A.; Bogo, D.; Pott, A. Fatty acid diets: Regulation of gut microbiota composition and obesity and its related metabolic dysbiosis. Int. J. Mol. Sci. 2020, 21, 4093. [Google Scholar] [CrossRef]
- Wilson, A.S.; Koller, K.R.; Ramaboli, M.C.; Nesengani, L.T.; Ocvirk, S.; Chen, C.; Thomas, T.K. Diet and the hu-man gut microbiome: An international review. Dig. Dis. Sci. 2020, 65, 723–740. [Google Scholar] [CrossRef] [Green Version]
- Amaretti, A.; Bottari, B.; Morreale, F.; Sardaro, M.L.S.; Angelino, D.; Raimondi, S.; Rossi, M.; Pellegrini, N. Potential prebiotic effect of a long-chain dextran produced by Weissella cibaria: An in vitro evaluation. Int. J. Food Sci. Nutr. 2020, 71, 563–571. [Google Scholar] [CrossRef]
- Thakkar, R.D.; Tuncil, Y.E.; Hamaker, B.R.; Lindemann, S.R. Maize bran particle size governs the community composition and metabolic output of human gut microbiota in in vitro fermentations. Front. Microbiol. 2020, 11, 1009. [Google Scholar] [CrossRef]
- Bas-Bellver, C.; Andrés, C.; Segui, L.; Barrera, C.; Jimenéz-Hernandéz, N.; Artacho, A.; Betoret, N.; Gosalbes, M.J. Valorization of Persimmon and Blueberry Byproducts to Obtain Functional Powders: In Vitro Digestion and Fermentation by Gut Microbiota. J. Agric. Food Chem. 2020, 68, 8080–8090. [Google Scholar] [CrossRef]
- Cheng, Y.; Wu, T.; Chu, X.; Tang, S.; Cao, W.; Liang, F.; Xu, X. Fermented blueberry pomace with antioxidant prop-erties improves fecal microbiota community structure and short chain fatty acids production in an in vitro mode. LWT 2020, 125, 109260. [Google Scholar] [CrossRef]
- Ji, X.G.; Chang, K.L.; Chen, M.; Zhu, L.L.; Osman, A.; Yin, H.; Zhao, L.M. In vitro fermentation of chitooligosaccha-rides and their effects on human fecal microbial community structure and metabolites. LWT 2021, 144, 111224. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, Y.; Chen, X.Q.; Aweya, J.J.; Cheong, K.L. Catabolism of Saccharina japonica polysaccharides and oligosaccharides by human fecal microbiota. LWT 2020, 130, 109635. [Google Scholar] [CrossRef]
- Liu, G.; Chen, H.; Chen, J.; Wang, X.; Gu, Q.; Yin, Y. Effects of bifidobacteria-produced exopolysaccharides on human gut microbiota in vitro. Appl. Microbiol. Biotechnol. 2018, 103, 1693–1702. [Google Scholar] [CrossRef] [PubMed]
- Navajas-Porras, B.; Pérez-Burillo, S.; Valverde-Moya, J.; Hinojosa-Nogueira, D.; Pastoriza, S.; Rufián-Henares, J. Effect of Cooking Methods on the Antioxidant Capacity of Plant Foods Submitted to in Vitro Digestion–Fermentation. Antioxidants 2020, 9, 1312. [Google Scholar] [CrossRef]
- Gong, L.; Cao, W.; Gao, J.; Wang, J.; Zhang, H.; Sun, B.; Yin, M. Whole Tibetan Hull-Less Barley exhibit stronger effect on promoting growth of genus Bifidobacterium than refined barley in vitro. J. Food Sci. 2018, 83, 1116–1124. [Google Scholar] [CrossRef]
- Vollmer, M.; Esders, S.; Farquharson, F.M.; Neugart, S.; Duncan, S.; Schreiner, M.; Louis, P.; Maul, R.; Rohn, S. Mutual Interaction of Phenolic Compounds and Microbiota: Metabolism of Complex Phenolic Apigenin-C- and Kaempferol-O-Derivatives by Human Fecal Samples. J. Agric. Food Chem. 2018, 66, 485–497. [Google Scholar] [CrossRef] [Green Version]
- Nie, C.; Yan, X.; Xie, X.; Zhang, Z.; Zhu, J.; Wang, Y.; Wang, X.; Xu, N.; Luo, Y.; Sa, Z.; et al. Structure of β-glucan from Tibetan hull-less barley and its in vitro fermentation by human gut microbiota. Chem. Biol. Technol. Agric. 2021, 8, 1–14. [Google Scholar] [CrossRef]
- Liu, Y.; Duan, X.; Duan, S.; Li, C.; Hu, B.; Liu, A.; Wu, Y.; Wu, H.; Chen, H.; Wu, W. Effects of in vitro digestion and fecal fermentation on the stability and metabolic behavior of polysaccharides from Craterellus cornucopioides. Food Funct. 2020, 11, 6899–6910. [Google Scholar] [CrossRef] [PubMed]
- Jayamanohar, J.; Devi, P.B.; Kavitake, D.; Priyadarisini, V.B.; Shetty, P.H. Prebiotic potential of water extractable polysaccharide from red kidney bean (Phaseolus vulgaris L.). LWT 2018, 101, 703–710. [Google Scholar] [CrossRef]
- Chen, G.; Xie, M.; Wan, P.; Chen, D.; Ye, H.; Chen, L.; Zeng, X.; Liu, Z. Digestion under saliva, simulated gastric and small intestinal conditions and fermentation in vitro by human intestinal microbiota of polysaccharides from Fuzhuan brick tea. Food Chem. 2018, 244, 331–339. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Chen, G.; Wan, P.; Hu, B.; Chen, L.; Ou, S.; Ye, H. Digestion under saliva, simulated gastric and small in-testinal conditions and fermentation in vitro of polysaccharides from the flowers of Camellia sinensis induced by hu-man gut microbiota. Food Funct. 2017, 8, 4619–4629. [Google Scholar] [CrossRef]
- Chen, C.; Huang, Q.; Fu, X.; Liu, R.H. In vitro fermentation of mulberry fruit polysaccharides by human fecal inocula and impact on microbiota. Food Funct. 2016, 7, 4637–4643. [Google Scholar] [CrossRef]
- Li, M.; Li, G.; Shang, Q.; Chen, X.; Liu, W.; Pi, X.; Zhu, L.; Yin, Y.; Yu, G.; Wang, X. In vitro fermentation of alginate and its derivatives by human gut microbiota. Anaerobe 2016, 39, 19–25. [Google Scholar] [CrossRef]
- Molino, S.; Lerma-Aguilera, A.; Jiménez-Hernández, N.; Gosalbes, M.J.; Rufián-Henares, J.Á.; Francino, M.P. Enrichment of food with tannin extracts promotes healthy changes in the human gut microbiota. Front. Microbiol. 2021, 12, 570. [Google Scholar] [CrossRef] [PubMed]
- Del juncal-Guzmán, D.; Hernández-Maldonado, L.M.; Sánchez-Burgos, J.A.; González-Aguilar, G.A.; Ruiz-Valdiviezo, V.M.; Tovar, J.; Sáyago-Ayerdi, S.G. In vitro gastrointestinal digestion and colonic fermentation of phenolic com-pounds in UV-C irradiated pineapple (Ananas comosus) snack-bars. LWT 2021, 138, 110636. [Google Scholar] [CrossRef]
- Gowd, V.; Bao, T.; Wang, L.; Huang, Y.; Chen, S.; Zheng, X.; Cui, S.; Chen, W. Antioxidant and antidiabetic activity of blackberry after gastrointestinal digestion and human gut microbiota fermentation. Food Chem. 2018, 269, 618–627. [Google Scholar] [CrossRef] [PubMed]
- Bresciani, L.; Dall’Asta, M.; Favari, C.; Calani, L.; Del Rio, D.; Brighenti, F. An in vitro exploratory study of dietary strategies based on polyphenol-rich beverages, fruit juices and oils to control trimethylamine production in the colon. Food Funct. 2018, 9, 6470–6483. [Google Scholar] [CrossRef]
- Navajas-Porras, B.; Pérez-Burillo, S.; Valverde-Moya, Á.; Hinojosa-Nogueira, D.; Pastoriza, S.; Rufián-Henares, J.Á. Effect of cooking methods on the antioxidant capacity of foods of animal origin submitted to in vitro digestion-fermentation. Antioxidants 2021, 10, 445. [Google Scholar] [CrossRef] [PubMed]
- Guan, N.; He, X.; Wang, S.; Liu, F.; Huang, Q.; Fu, X.; Chen, T.; Zhang, B. Cell Wall Integrity of Pulse Modulates the in Vitro Fecal Fermentation Rate and Microbiota Composition. J. Agric. Food Chem. 2019, 68, 1091–1100. [Google Scholar] [CrossRef] [PubMed]
- Nsor-Atindana, J.; Zhou, Y.X.; Saqib, N.; Chen, M.; Goff, H.D.; Ma, J.; Zhong, F. Enhancing the prebiotic effect of cellulose biopolymer in the gut by physical structuring via particle size manipulation. Food Res. Int. 2019, 131, 108935. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Burillo, S.; Pastoriza, S.; Jiménez-Hernández, N.; D’Auria, G.; Francino, M.P.; Rufián-Henares, J.A. Effect of Food Thermal Processing on the Composition of the Gut Microbiota. J. Agric. Food Chem. 2018, 66, 11500–11509. [Google Scholar] [CrossRef]
- Brahma, S.; Weier, S.A.; Rose, D.J. Moisture content during extrusion of oats impacts the initial fermentation metabolites and probiotic bacteria during extended fermentation by human fecal microbiota. Food Res. Int. 2017, 97, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Arcila, J.A.; Weier, S.A.; Rose, D.J. Changes in dietary fiber fractions and gut microbial fermentation properties of wheat bran after extrusion and bread making. Food Res. Int. 2015, 74, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Chen, Y.A.; Tuohy, K.M. A comparative in vitro investigation into the effects of cooked meats on the hu-man faecal microbiota. Anaerobe 2010, 16, 572–577. [Google Scholar] [CrossRef]
- Singh, V.; Hwang, N.; Ko, G.; Tatsuya, U. Effects of digested Cheonggukjang on human microbiota assessed by in vitro fecal fermentation. J. Microbiol. 2021, 59, 217–227. [Google Scholar] [CrossRef]
- Young, W.; Arojju, S.K.; McNeill, M.R.; Rettedal, E.; Gathercole, J.; Bell, N.; Payne, P. Feeding Bugs to Bugs: Edible Insects Modify the Human Gut Microbiome in an in vitro Fermentation Model. Front. Microbiol. 2020, 11, 1763. [Google Scholar] [CrossRef] [PubMed]
- Miclotte, L.; De Paepe, K.; Rymenans, L.; Callewaert, C.; Raes, J.; Rajkovic, A.; Van Camp, J.; Van de Wiele, T. Dietary Emulsifiers Alter Composition and Activity of the Human Gut Microbiota in vitro, Irrespective of Chemical or Natural Emulsifier Origin. Front. Microbiol. 2020, 11, 577474. [Google Scholar] [CrossRef]
- Dou, Z.; Chen, C.; Fu, X. Bioaccessibility, antioxidant activity and modulation effect on gut microbiota of bioactive compounds from Moringa oleifera Lam. leaves during digestion and fermentation in vitro. Food Funct. 2019, 10, 5070–5079. [Google Scholar] [CrossRef]
- López-Barrera, D.M.; Vázquez-Sánchez, K.; Loarca-Piña, M.G.F.; Campos-Vega, R. Spent coffee grounds, an innovative source of colonic fermentable compounds, inhibit inflammatory mediators in vitro. Food Chem. 2016, 212, 282–290. [Google Scholar] [CrossRef]
- Mandalari, G.; Chessa, S.; Bisignano, C.; Chan, L.; Carughi, A. The effect of sun-dried raisins (Vitis vinifera L.) on the in vitro composition of the gut microbiota. Food Funct. 2016, 7, 4048–4060. [Google Scholar] [CrossRef]
- Krishnamoorthy, R.; Adisa, A.R.; Periasamy, V.S.; Athinarayanan, J.; Pandurangan, S.-B.; Alshatwi, A.A. Colonic Bacteria-Transformed Catechin Metabolite Response to Cytokine Production by Human Peripheral Blood Mononuclear Cells. Biomolecules 2019, 9, 830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nissen, L.; Casciano, F.; Gianotti, A. Intestinal fermentation in vitro models to study food-induced gut microbiota shift: An updated review. FEMS Microbiol. Lett. 2020, 367, fnaa097. [Google Scholar] [CrossRef]
- Sáyago-Ayerdi, S.G.; Venema, K.; Tabernero, M.; Sarriá, B.; Bravo, L.L.; Mateos, R. Bioconversion by gut microbiota of predigested mango (Mangifera indica L.) ‘Ataulfo’ peel polyphenols assessed in a dynamic (TIM-2) in vitro model of the human colon. Food Res. Int. 2020, 139, 109963. [Google Scholar] [CrossRef]
- Marzorati, M.; Abbeele, P.V.D.; Bubeck, S.S.; Bayne, T.; Krishnan, K.; Young, A.; Mehta, D.; DeSouza, A. Bacillus subtilis HU58 and Bacillus coagulans SC208 Probiotics Reduced the Effects of Antibiotic-Induced Gut Microbiome Dysbiosis in An M-SHIME® Model. Microorganisms 2020, 8, 1028. [Google Scholar] [CrossRef] [PubMed]
- Poeker, S.A.; Geirnaert, A.; Berchtold, L.; Greppi, A.; Krych, L.; Steinert, R.E.; Lacroix, C. Understanding the prebi-otic potential of different dietary fibers using an in vitro continuous adult fermentation model (PolyFermS). Sci. Rep. 2018, 8, 4318. [Google Scholar] [CrossRef] [PubMed]
- Casarotti, S.N.; Borgonovi, T.F.; Tieghi, T.D.M.; Sivieri, K.; Penna, A.L.B. Probiotic low-fat fermented goat milk with passion fruit by-product: In vitro effect on obese individuals’ microbiota and on metabolites production. Food Res. Int. 2020, 136, 109453. [Google Scholar] [CrossRef] [PubMed]
- Gil-Sánchez, I.; Cueva, C.; Tamargo, A.; Quintela, J.C.; de la Fuente, E.; Walker, A.; Moreno-Arribas, M.V.; Bartolomé, B. Application of the dynamic gastrointestinal simulator (simgi®) to assess the impact of probiotic supplementation in the metabolism of grape polyphenols. Food Res. Int. 2019, 129, 108790. [Google Scholar] [CrossRef]
- Bianchi, F.; Larsen, N.; Tieghi, T.D.M.; Adorno, M.A.T.; Kot, W.; Saad, S.; Jespersen, L.; Sivieri, K. Modulation of gut microbiota from obese individuals by in vitro fermentation of citrus pectin in combination with Bifidobacterium longum BB-46. Appl. Microbiol. Biotechnol. 2018, 102, 8827–8840. [Google Scholar] [CrossRef] [Green Version]
- Patrignani, F.; Parolin, C.; D’Alessandro, M.; Siroli, L.; Vitali, B.; Lanciotti, R. Evaluation of the fate of Lactobacillus crispatus BC4, carried in Squacquerone cheese, throughout the simulator of the human intestinal microbial ecosystem (SHIME). Food Res. Int. 2020, 137, 109580. [Google Scholar] [CrossRef] [PubMed]
- Tamargo, A.; Cueva, C.; Alvarez, M.D.; Herranz, B.; Moreno-Arribas, M.V.; Laguna, L. Physical effects of dietary fibre on simulated luminal flow, studied by in vitro dynamic gastrointestinal digestion and fermentation. Food Funct. 2019, 10, 3452–3465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van den Abbeele, P.; Marzorati, M.; Derde, M.; De Weirdt, R.; Joan, V.; Possemiers, S.; Van de Wiele, T. Arabinoxylans, inulin and Lactobacillus reuteri 1063 repress the adherent-invasive Escherichia coli from mucus in a muco-sa-comprising gut model. NPJ Biofilms Microbiomes 2016, 2, 1–8. [Google Scholar] [CrossRef]
- Barroso, E.; Van De Wiele, T.; Jiménez-Girón, A.; Muñoz-Gonzalez, I.; Martín-Alvarez, P.J.; Moreno-Arribas, M.V.; Bartolomé, B.; Pelaez, C.; Martinez-Cuesta, M.C.; Requena, T. Lactobacillus plantarum IFPL935 impacts colonic metabolism in a simulator of the human gut microbiota during feeding with red wine polyphenols. Appl. Microbiol. Biotechnol. 2014, 98, 6805–6815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dupont, D.; Alric, M.; Blanquet-Diot, S.; Bornhorst, G.; Cueva, C.; Deglaire, A.; Mackie, A.R. Can dynamic in vitro digestion systems mimic the physiological reality? Crit. Rev. Food Sci. Nutr. 2019, 59, 1546–1562. [Google Scholar] [CrossRef] [Green Version]
- Venema, K.; Verhoeven, J.; Verbruggen, S.; Keller, D. Xylo-oligosaccharides from sugarcane show prebiotic potential in a dynamic computer-controlled in vitro model of the adult human large intestine. Benef. Microbes 2020, 11, 191–200. [Google Scholar] [CrossRef]
- Cárdenas-Castro, A.P.; Bianchi, F.; Tallarico-Adorno, M.A.; Montalvo-González, E.; Sáyago-Ayerdi, S.G.; Sivieri, K. In vitro colonic fermentation of Mexican “taco” from corn-tortilla and black beans in a Simulator of Human Microbial Ecosystem (SHIME®) system. Food Res. Int. 2019, 118, 81–88. [Google Scholar] [CrossRef] [Green Version]
- Chassaing, B.; Van De Wiele, T.; De Bodt, J.; Marzorati, M.; Gewirtz, A.T. Dietary emulsifiers directly alter human microbiota composition and gene expression ex vivo potentiating intestinal inflammation. Gut 2017, 66, 1414–1427. [Google Scholar] [CrossRef]
- Fehlbaum, S.; Chassard, C.; Haug, M.C.; Fourmestraux, C.; Derrien, M.; Lacroix, C. Design and investigation of PolyFermS in vitro continuous fermentation models inoculated with immobilized fecal microbiota mimicking the el-derly colon. PLoS ONE 2015, 10, e0142793. [Google Scholar] [CrossRef] [PubMed]
- Berner, A.Z.; Fuentes, S.; Dostal, A.; Payne, A.N.; Gutierrez, P.V.; Chassard, C.; Grattepanche, F.; De Vos, W.M.; Lacroix, C. Novel Polyfermentor Intestinal Model (PolyFermS) for Controlled Ecological Studies: Validation and Effect of pH. PLoS ONE 2013, 8, e77772. [Google Scholar] [CrossRef] [Green Version]
- Salgaço, M.K.; Perina, N.P.; Tomé, T.M.; Mosquera, E.M.B.; Lazarini, T.; Sartoratto, A.; Sivieri, K. Probiotic infant cereal improves children’s gut microbiota: Insights using the Simulator of Human Intestinal Microbial Ecosystem (SHIME®). Food Res. Int. 2021, 143, 110292. [Google Scholar] [CrossRef] [PubMed]
- Oddi, S.; Huber, P.; Duque, A.R.F.; Vinderola, G.; Sivieri, K. Breast-milk derived potential probiotics as strategy for the management of childhood obesity. Food Res. Int. 2020, 137, 109673. [Google Scholar] [CrossRef]
- Roselino, M.N.; Sakamoto, I.K.; Adorno, M.A.T.; Canaan, J.M.M.; de Valdez, G.F.; Rossi, E.A.; Sivieri, K.; Cavallini, D.C.U. Effect of fermented sausages with probiotic Enterococcus faecium CRL 183 on gut microbiota using dynamic colonic model. LWT 2020, 132, 109876. [Google Scholar] [CrossRef]
- Worametrachanon, S.; Apichartsrangkoon, A.; Chaikham, P.; Abbeele, P.V.D.; Van De Wiele, T.; Wirjantoro, T.I. Effect of encapsulated Lactobacillus casei 01 along with pressurized-purple-rice drinks on colonizing the colon in the digestive model. Appl. Microbiol. Biotechnol. 2014, 98, 5241–5250. [Google Scholar] [CrossRef]
- Chaikham, P.; Apichartsrangkoon, A. Effects of encapsulated Lactobacillus acidophilus along with pasteurized lon-gan juice on the colon microbiota residing in a dynamic simulator of the human intestinal microbial ecosystem. Appl. Microbiol. Biotechnol. 2014, 98, 485–495. [Google Scholar] [CrossRef]
- Sivieri, K.; Morales, M.L.V.; Adorno, M.A.T.; Sakamoto, I.K.; Saad, S.M.I.; Rossi, E.A. Lactobacillus acidophilus CRL 1014 improved “gut health” in the SHIME® reactor. BMC Gastroenterol. 2013, 13, 1–9. [Google Scholar] [CrossRef]
- Possemiers, S.; Marzorati, M.; Verstraete, W.; Van De Wiele, T. Bacteria and chocolate: A successful combination for probiotic delivery. Int. J. Food Microbiol. 2010, 141, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Giuliani, C.; Marzorati, M.; Innocenti, M.; Vilchez-Vargas, R.; Vital, M.; Pieper, D.H.; Mulinacci, N. Dietary sup-plement based on stilbenes: A focus on gut microbial metabolism by the in vitro simulator M-SHIME®. Food Funct. 2016, 7, 4564–4575. [Google Scholar] [CrossRef]
- Kemperman, R.A.; Gross, G.; Mondot, S.; Possemiers, S.; Marzorati, M.; Van de Wiele, T.; Doré, J.; Vaughan, E.E. Impact of polyphenols from black tea and red wine/grape juice on a gut model microbiome. Food Res. Int. 2013, 53, 659–669. [Google Scholar] [CrossRef]
- Allsopp, P.; Possemiers, S.; Campbell, D.; Oyarzábal, I.S.; Gill, C.; Rowland, I. An exploratory study into the putative prebiotic activity of fructans isolated from Agave angustifolia and the associated anticancer activity. Anaerobe 2013, 22, 38–44. [Google Scholar] [CrossRef]
- Terpend, K.; Possemiers, S.; Daguet, D.; Marzorati, M. Arabinogalactan and fructo-oligosaccharides have a different fermentation profile in the Simulator of the Human Intestinal Microbial Ecosystem (SHIME®). Environ. Microbiol. Rep. 2013, 5, 595–603. [Google Scholar] [CrossRef] [PubMed]
- Marzorati, M.; Verhelst, A.; Luta, G.; Sinnott, R.; Verstraete, W.; Van De Wiele, T.; Possemiers, S. In vitro modulation of the human gastrointestinal microbial community by plant-derived polysaccharide-rich dietary supplements. Int. J. Food Microbiol. 2010, 139, 168–176. [Google Scholar] [CrossRef]
- Bondue, P.; Lebrun, S.; Taminiau, B.; Everaert, N.; LaPointe, G.; Hendrick, C.; Delcenserie, V. Effect of Bifidobacte-rium crudilactis and 3′-sialyllactose on the toddler microbiota using the SHIME® model. Food Res. Int. 2020, 138, 109755. [Google Scholar] [CrossRef] [PubMed]
- Freire, F.C.; Adorno, M.A.T.; Sakamoto, I.K.; Antoniassi, R.; Chaves, A.C.S.D.; dos Santos, K.M.O.; Sivieri, K. Impact of multi-functional fermented goat milk beverage on gut microbiota in a dynamic colon model. Food Res. Int. 2017, 99, 315–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bianchi, F.; Rossi, E.A.; Sakamoto, I.K.; Adorno, M.A.T.; Van de Wiele, T.; Sivieri, K. Beneficial effects of fermented vegetal beverages on human gastrointestinal microbial ecosystem in a simulator. Food Res. Int. 2014, 64, 43–52. [Google Scholar] [CrossRef] [Green Version]
- Marzorati, M.; Vilchez-Vargas, R.; Bussche, J.V.; Truchado, P.; Jauregui, R.; El Hage, R.A.; Pieper, D.H.; Vanhaecke, L.; Van De Wiele, T. High-fiber and high-protein diets shape different gut microbial communities, which ecologically behave similarly under stress conditions, as shown in a gastrointestinal simulator. Mol. Nutr. Food Res. 2016, 61, 1600150. [Google Scholar] [CrossRef] [Green Version]
- Duque, A.L.R.F.; Monteiro, M.; Adorno, M.A.T.; Sakamoto, I.K.; Sivieri, K. An exploratory study on the influence of orange juice on gut microbiota using a dynamic colonic model. Food Res. Int. 2016, 84, 160–169. [Google Scholar] [CrossRef]
- Sáyago-Ayerdi, S.G.; Zamora-Gasga, V.M.; Venema, K. Changes in gut microbiota in predigested Hibiscus sabdar-iffa L calyces and Agave (Agave tequilana weber) fructans assessed in a dynamic in vitro model (TIM-2) of the human colon. Food Res. Int. 2020, 132, 109036. [Google Scholar] [CrossRef]
- Míguez, B.; Vila, C.; Venema, K.; Parajó, J.C.; Alonso, J.L. Potential of high-and low-acetylated galactoglucoman-nooligosaccharides as modulators of the microbiota composition and their activity: A comparison using the in vitro model of the human colon TIM-2. J. Agric. Food Chem. 2020, 68, 7617–7629. [Google Scholar] [CrossRef]
- Larsen, N.; de Souza, C.B.; Krych, L.; Cahú, T.B.; Wiese, M.; Kot, W.; Hansen, K.M.; Blennow, A.; Venema, K.; Jespersen, L. Potential of Pectins to Beneficially Modulate the Gut Microbiota Depends on Their Structural Properties. Front. Microbiol. 2019, 10, 223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramasamy, U.S.; Venema, K.; Gruppen, H.; Schols, H.A. The fate of chicory root pulp polysaccharides during fermentation in the TNO in vitro model of the colon (TIM-2). Bioact. Carbohydr. Diet. Fibre 2014, 4, 48–57. [Google Scholar] [CrossRef]
- Cárdenas-Castro, A.P.; Venema, K.; Sarriá, B.; Bravo, L.; Sáyago-Ayerdi, S.G.; Mateos, R. Study of the impact of a dynamic in vitro model of the colon (TIM-2) in the phenolic composition of two Mexican sauces. Food Res. Int. 2021, 139, 109917. [Google Scholar] [CrossRef]
- Martina, A.; Felis, G.; Corradi, M.; Maffeis, C.; Torriani, S.; Venema, K. Effects of functional pasta ingredients on different gut microbiota as revealed by TIM-2 in vitro model of the proximal colon. Benef. Microbes 2019, 10, 301–313. [Google Scholar] [CrossRef]
- Kortman, G.A.; Dutilh, B.E.; Maathuis, A.J.; Engelke, U.F.; Boekhorst, J.; Keegan, K.P.; Tjalsma, H. Microbial me-tabolism shifts towards an adverse profile with supplementary iron in the TIM-2 in vitro model of the human colon. Front. Microbiol. 2016, 6, 1481. [Google Scholar] [CrossRef] [Green Version]
- Gil-Sánchez, I.; Cueva, C.; Sanz-Buenhombre, M.; Guadarrama, A.; Moreno-Arribas, M.V.; Bartolomé, B. Dynamic gastrointestinal digestion of grape pomace extracts: Bioaccessible phenolic metabolites and impact on human gut microbiota. J. Food Compos. Anal. 2018, 68, 41–52. [Google Scholar] [CrossRef]
- Cueva, C.; Jiménez-Girón, A.; Muñoz-Gonzalez, I.; Esteban-Fernández, A.; Gil-Sánchez, I.; Dueñas, M.; Martín-Álvarez, P.; Pozo-Bayón, M.; Bartolomé, B.; Moreno-Arribas, M. Application of a new Dynamic Gastrointestinal Simulator (SIMGI) to study the impact of red wine in colonic metabolism. Food Res. Int. 2015, 72, 149–159. [Google Scholar] [CrossRef]
- Dostal, A.; Lacroix, C.; Bircher, L.; Follador, R.; Zimmermann, M.B.; Chassard, C. Iron modulates butyrate production by a child gut microbiota in vitro. mBio 2015, 6, e01453-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Mantrana, I.; Selma-Royo, M.; Alcantara, C.; Collado, M.C. Shifts on gut microbiota associated to mediterranean diet adherence and specific dietary intakes on general adult population. Front. Microbiol. 2018, 9, 890. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, G.; Peng, Y.; Rui, Y.; Zeng, X.; Ye, H. Simulated digestion and fermentation in vitro with human gut microbiota of polysaccharides from Coralline pilulifera. LWT 2019, 100, 167–174. [Google Scholar] [CrossRef]
- Sergaki, C.; Lagunas, B.; Lidbury, I.; Gifford, M.L.; Schäfer, P. Challenges and Approaches in Microbiome Research: From Fundamental to Applied. Front. Plant Sci. 2018, 9, 1205. [Google Scholar] [CrossRef]
- Zhang, X.; Li, L.; Butcher, J.; Stintzi, A.; Figeys, D. Advancing functional and translational microbiome research using meta-omics approaches. Microbiome 2019, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zamora-Gasga, V.M.; Loarca-Piña, G.; Vázquez-Landaverde, P.A.; Ortiz-Basurto, R.I.; Tovar, J.; Sáyago-Ayerdi, S.G. In vitro colonic fermentation of food ingredients isolated from Agave tequilana Weber var. azul applied on granola bars. LWT 2015, 60, 766–772. [Google Scholar] [CrossRef]
- Gong, L.; Chi, J.; Zhang, Y.; Wang, J.; Sun, B. In vitro evaluation of the bioaccessibility of phenolic acids in different whole wheats as potential prebiotics. LWT 2018, 100, 435–443. [Google Scholar] [CrossRef]
- Pérez-Burillo, S.; Mehta, T.; Pastoriza, S.; Kramer, D.L.; Paliy, O.; Rufián-Henares, J. Potential probiotic salami with dietary fiber modulates antioxidant capacity, short chain fatty acid production and gut microbiota community structure. LWT 2019, 105, 355–362. [Google Scholar] [CrossRef]
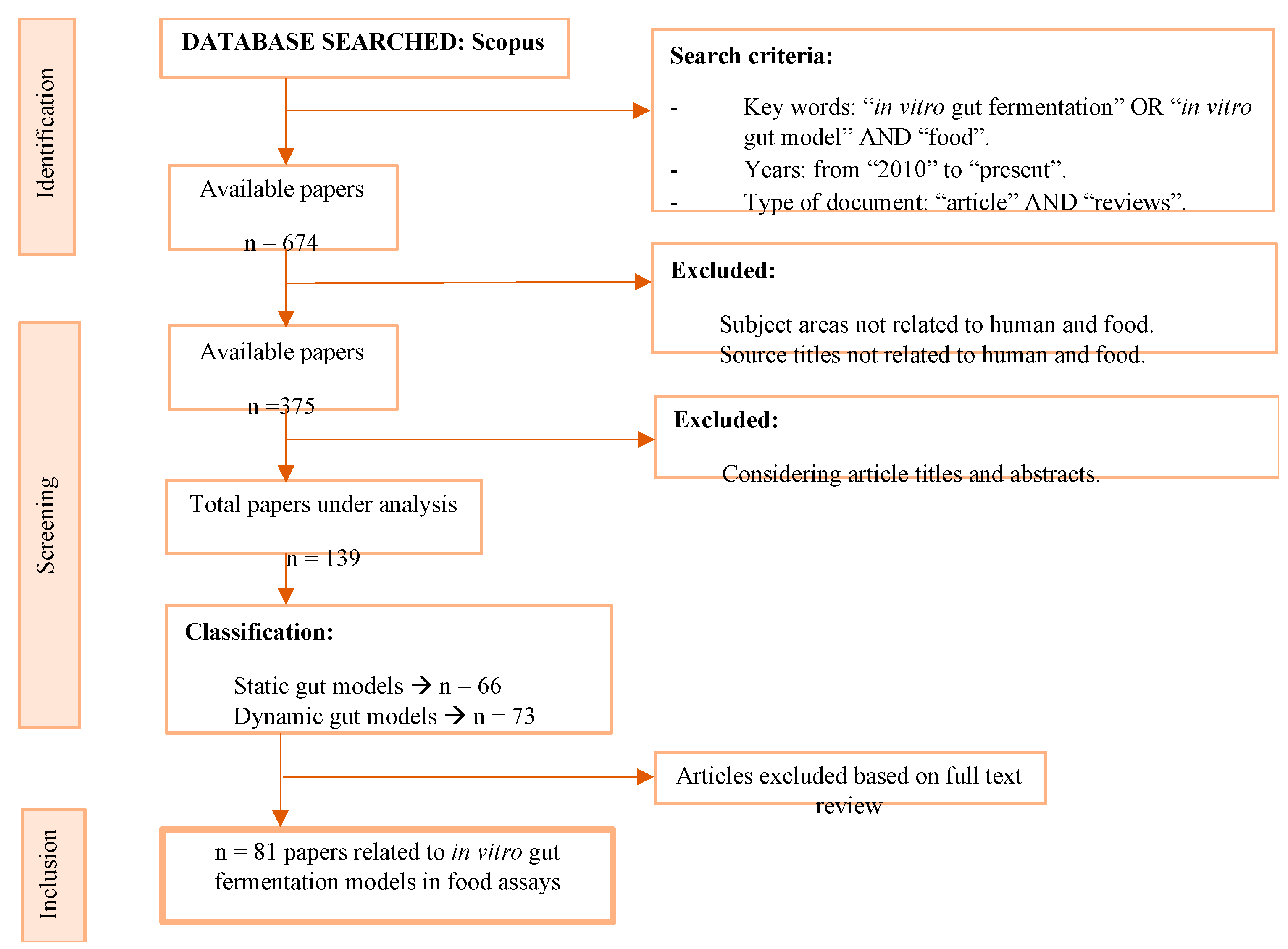
| Main Objective | Study Subject | References |
|---|---|---|
| To assay the prebiotic effect of polysaccharides and oligosaccharides on gut microbiota and their metabolites | Polysaccharides and oligosaccharides from: -Fruits: Mulberry fruit. -Fungi: Craterellus cornucopioides. -Algae: Saccharina japonica, and Corallina pilulifera. -Plants: flowers of Camellia sinensis and A. tequilana Weber var. -Grains: Tibetan hull-less barley. -Bacteria: Weissella cibaria and Bifidobacteria. -Beans: Red kidney bean. -Tea: Fuzhuan brick tea. | [30,31,32,36,37,38,39,40,41,42] |
| To assay the prebiotic effect of some polyphenols on gut microbiota and their metabolites. | Different polyphenols from: -Foods: cereal, meat, and dairy. -Drinks: coffee, tea, wine, and different juices. -New snacks: Pineapple snack bars. -Industrial residues: Persimmon and blueberry residues. -Pomaces: Blueberry pomace. | [28,29,34,43,44,45,46] |
| To assay the effect of sample processing on gastrointestinal digestion and fermentation, gut microbiota, and their metabolites. | -Dairy, egg, fish, and meat products after boiling, frying, grilling, and roasting. -Plant food products (alcoholic drinks, cereals, cocoa, coffee, fruits, legumes, nuts, oils, tubers, and vegetables) after boiling, frying, grilling, roasting, and toasting. -Enzymatic modification of the cell wall integrity: cotyledon cells from pinto bean seeds. -Extrusion: whole grain oats and wheat bran. -Particle size manipulation: maize bran, cellulose, and whole grain oat flakes. | [27,33,47,48,49,50,51,52,53] |
| To assay the effect of whole foods or additives on gut microbiota and their metabolites. | -Cheonggukjang: a Korean traditional fermented soybean soup. -Edible insects: black field cricket nymphs, grass grub larvae, and wax moth larvae. -Moringa oleifera Lam. Leaves -Probiotic salami. -Tibetan hull-less barley and refined Tibetan hull-less barley. -Spent coffee grounds. -Sun-dried raisins. Additives such as: -Dietary emulsifiers: sodium CMC, P80, soy lecithin, sophorolipids, and rhamnolipids. | [34,50,54,55,56,57,58,59] |
| To assay the gut microbiota metabolism in response to food components. | Biotransformation of polyphenols by human gut microbiota: -Catechin. -Flavonoid O- and C-glycosides. -[6]-Shogaol from ginger. | [35,60] |
| Dynamic Model | Main Objective and Examples | References |
|---|---|---|
| SHIME | Assessing the probiotic effect of some bacteria: -Bacteria (e.g., Lactobacillus, Bifidobacterium, or Enterococcus) carried in cheese, chocolate, cereals, sausages, and drinks. -Probiotic formulations (e.g., Lactobacillus, Bifidobacterium, or Bacillus) | [63,68,78,79,79,80,81,82,83,84] |
| Assessing the prebiotic effect of some compounds: -Polyphenols (e.g., black tea or red wine grape extract, t-resveratrol, and ε-viniferin extract). -Polysaccharides (e.g., branched fructans, arabinogalactan, fructooligosaccharides, commercially available plant polysaccharide) | [85,86,87,88,89] | |
| Assessing the symbiotic effect (probiotic + prebiotic): -Milks and beverages fermented by different microorganisms (e.g., Lactobacillus) and supplemented with different compounds (e.g., passion fruit, grape pomace) -Probiotic and prebiotic formulations (e.g., Bifidobacterium + 3′-sialyllactose, Lactobacillus + red win polyphenolic extract, Lactobacillus + fructooligosaccharides, Bifidobacterium + pectins -Repressing Escherichia coli colonization of the gut mucus. | [65,67,70,71,90,91,92] | |
| Other applications assessing the effect of: -Food ingredients (e.g., Mexican “taco” from corn tortilla and black beans). -Dietary emulsifiers (e.g., polysorbate 80 and carboxymethylcellulose). -Antibiotics (e.g., amoxicillin, ciprofloxacin, and tetracycline). -Processing (e.g., fresh and pasteurized orange juice). | [75,93,94] | |
| TIM-2 | Assessing the prebiotic effect of some compounds: -Polyphenols (e.g., polyphenols in pre-digested mango peel, pre-digested Hibiscus sabdariffa calyces) -Polysaccharides: (e.g., High- and low-acetylated galactoglucomannooligosaccharides, agave fructans, xylo-oligosaccharides, pectins, chicory root pulp polysaccharides) | [62,73,95,96,97,98] |
| Other applications assessing the effect of: -Supplementation with minerals (e.g., iron). -Food ingredients (e.g., two types of Mexican sauces) -Symbiotic effect (e.g., functional pasta with Bacillus and β-glucans). | [99,100,101] | |
| SIMGI | Assessing the probiotic effect of some bacteria -Lactobacillus. | [66] |
| Assessing the prebiotic effect of some compounds: -Polysaccharides: (e.g., apple, potato, oat and psyllium fibres, hydroxypropyl methylcellulose, and microcrystalline cellulose). -Polyphenols: (e.g., grape pomace extracts). | [69,102] | |
| Other applications assessing the effect of: -Food (e.g., red wine). | [103] | |
| PolyFermS | Assessing the prebiotic effect of some compounds: -Polysaccharides: (e.g., fructo-oligosaccharides, β-glucan, α-galactooligosaccharide, xylo-oligosaccharide | [64] |
| Other uses assessing the effect of: -Supplementation with minerals (e.g., iron). | [104] |
| Main Objective | Analytical Determinations | References |
|---|---|---|
| To assay the prebiotic effect of different polysaccharides or oligosaccharides on gut microbiota and their metabolites. | -Microbiota analysis: 16S rDNA Sequencing, qPCR, plate counting with selective media or PCR-DGGE gel. -SCFA analysis: GC-FID. -SCFA and BCFA analysis: ion-chromatography. -Metabolite analysis in fermentation broth: GC-MS or HPLC. -pH measurement: pH meter. -Ammonia: anion measurer connected to a selective-ion electrode. -Volume gas production: graduated syringe displacement method. -Other analysis: Reducing sugar (DNS method), viscosity measurements (rheometer), or particle size measurements (laser diffraction particle size analyser). | [26,30,31,36,37,38,42,54,64,69,79,80,97,109] |
| To assay the prebiotic effect of different polyphenols on gut microbiota and their metabolites. | -Microbiota analysis: 16S rDNA Sequencing, plate counting with selective media, qPCR or PCR-DGGE gel. -SCFA and metabolite analysis: GC-FID, HPLC or GC-MS. -Phenolic compounds analysis: HPLC-MS, UPLC-MS, UHPLC-MS, or UPLC-ESI–MS/MS. -Ammonium: pH/ion meter coupled to an ammonium selective-ion electrode. | [29,43,44,46,85,86,95,99,102] |
| To assay the effect of sample processing on antioxidant capacity during gastrointestinal digestion and fermentation. | -Antioxidant tests: TEACDPPH assay, TEACFRAP assay, Folin–Ciocalteu assay and TEACABTS assay. -Determination of phenolic acids: LC-MS or HPLC. -Microbiota analysis: 16S rDNA Sequencing. | [33,45,47,110] |
| To assay the effect of dietary emulsifiers on gut microbiota and its pro-inflammatory contributions. | -SCFA analysis: GC-FID. -Microbiota analysis: 16S rDNA Sequencing and metatranscriptomic analysis. -Other analysis: Cell culture for flagellin detection (flagellin is a virulence factor). | [56,75] |
| To assay sample processing (extrusion, and diverse thermal treatments) on gut microbiota | -Gut microbiota contents: 16S rDNA Sequencing, qPCR, or FISH. -SCFA analysis: HPLC or GC. | [50,51,53] |
| To assay functional foods effect on gut microbiota. | -Antioxidant assays: TEACABTS assay, TEACFRAP assay, TEACOH method, TEACAAPH method, GEACRED method, solid residue antioxidant capacity, Folin–Ciocalteu colorimetric method. -Analysis of phenolic acids: HPLC. -SCFA analysis: HPLC or GC-FID. -Microbiota analysis: 16S rDNA sequencing or plate counting with selective media. | [34,59,111] |
| To assay the probiotic effect of some bacteria on gut microbiota. | -SCFA analysis: GC-FID. -pH measurement: pH meter. -Ammonium: Using a selective ion meter coupled to a selective-ion electrode or realising ammonium as ammonia and titrating it with HCl. -Microbiota analysis: qPCR, plate counting, PCR-DGGE, or 16S rRNA sequencing. | [68,79,81,83] |
| To assay the symbiotic (prebiotic + probiotic) on gut microbiota | -Microbiota analysis: 16S rDNA sequencing, plate counts, PCR-DGGE, or qPCR. -SCFA analysis: GC-FID. -Ammonium analysis: Using an anion measurer connected to a selective-ion electrode or realising ammonium as ammonia and titrating it with HCl. -Lactic acid: spectrophotometrically with an enzymatic D-/L-lactic acid Kit. -Phenolic metabolites analysis: UPLC-MS/MS. | [65,67,71,84,92] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Veintimilla-Gozalbo, E.; Asensio-Grau, A.; Calvo-Lerma, J.; Heredia, A.; Andrés, A. In Vitro Simulation of Human Colonic Fermentation: A Practical Approach towards Models’ Design and Analytical Tools. Appl. Sci. 2021, 11, 8135. https://doi.org/10.3390/app11178135
Veintimilla-Gozalbo E, Asensio-Grau A, Calvo-Lerma J, Heredia A, Andrés A. In Vitro Simulation of Human Colonic Fermentation: A Practical Approach towards Models’ Design and Analytical Tools. Applied Sciences. 2021; 11(17):8135. https://doi.org/10.3390/app11178135
Chicago/Turabian StyleVeintimilla-Gozalbo, Elena, Andrea Asensio-Grau, Joaquim Calvo-Lerma, Ana Heredia, and Ana Andrés. 2021. "In Vitro Simulation of Human Colonic Fermentation: A Practical Approach towards Models’ Design and Analytical Tools" Applied Sciences 11, no. 17: 8135. https://doi.org/10.3390/app11178135
APA StyleVeintimilla-Gozalbo, E., Asensio-Grau, A., Calvo-Lerma, J., Heredia, A., & Andrés, A. (2021). In Vitro Simulation of Human Colonic Fermentation: A Practical Approach towards Models’ Design and Analytical Tools. Applied Sciences, 11(17), 8135. https://doi.org/10.3390/app11178135







