A Randomized Bag-of-Birds Approach to Study Robustness of Automated Audio Based Bird Species Classification
Abstract
:1. Introduction
2. Methods
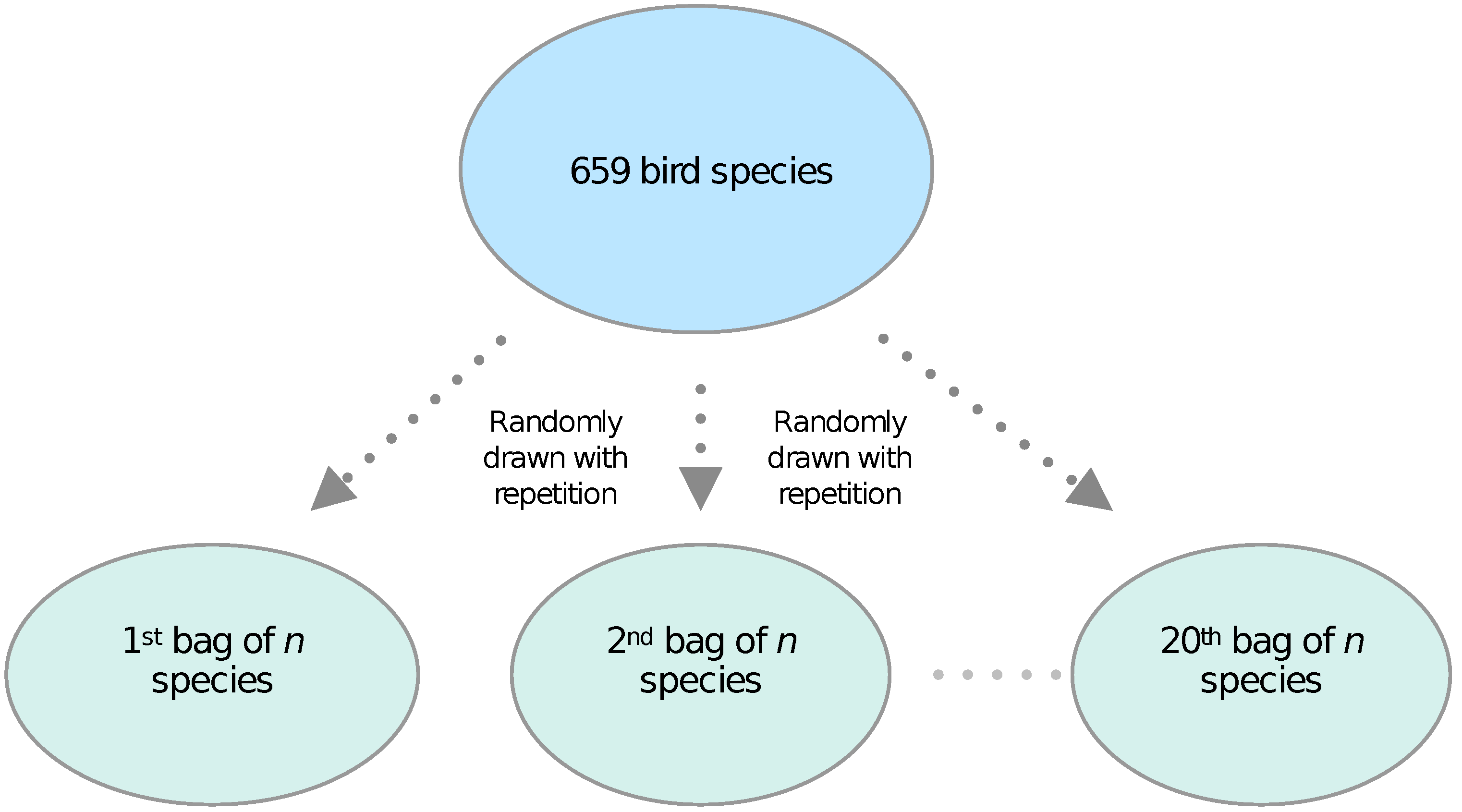
2.1. Bags-of-Birds Approach: Performing Randomized Classification Experiments
2.2. Feature Extraction
- Spectral Centroid: The spectral centroid measures the frequency where the energy of a spectrum is centered. In other words, it localizes the center of mass of the spectrum and is calculated as a weighted mean of the frequencies that the signal is composed of:where is the spectral magnitude at frequency bin k, and represents the center frequency of the bin [23].
- Spectral Rolloff: The spectral rolloff gives the frequency , below which, a pre-defined percentage (usually set to 85%) of the total spectral energy is concentrated [24].
- Zero-Crossing Rate: The zero-crossing rate r measures the smoothness of a signal. It is the rate at which a signal changes its sign from negative to positive or vice versa [25].
- The root-mean-square energy (RMSE): The root-mean-square energy of a signal gives the signal’s total energy and is defined as:where is the spectral magnitude at frequency bin k [17].
- Spectral Bandwith: measures if the power spectrum is concentrated around the spectral centroid or spread across the spectrum. It is computed as:where is the spectral centroid, is the spectral magnitude at frequency bin k [26].
- Mel-Frequency Cepstral Coefficients (MFCCs): Mel-Frequency Cepstral Coefficients are inspired by human auditory perception. After computing the Fourier transform of a signal, the magnitude spectrum is projected to the Mel scale, which emphasizes relevant frequencies in a non-linear way—a small bandwidth at low frequencies and large bandwidth at high frequencies. The Mel scale approximates human auditory response better than linearly spaced frequency bands. The output is log transformed, and MFCCs are obtained by taking a discrete cosine transform of the logarithmic outputs [27]. In this contribution, the analysis frames are split into windows of lengths 512 and compute the first 20 MFCC values as features in our system.
2.3. Classification Model
2.4. Measuring Classification Success
| the number of classifications that are true positives for species j, | |
| the number of classifications that are true negatives for species j, | |
| the number of classifications that are false positives for species j, and | |
| the number of classifications that are false negatives for species j. |
- Precision: This metric gives the measure of reliability of our predictions. The formula to compute the precision for a bird species j isTherefore, the precision for a species j indicates how many true positives the model predicted out of all positives. Therefore, the higher the precision, the more confident a model is about its predictions. In order to compute the precision for the entire test dataset, we average over all species
- Recall: This metric gives the measure of predictive power of a model. The formula to compute recall for each bird species j isThus, to recall, for a class species j, would indicate, of all actual positives in the test dataset, how many did the model predict as positive. Therefore, the higher the recall, the more positive samples the model correctly classified as positive. In order to compute the recall for the entire test dataset, we averaged over all species
- Accuracy: While precision and recall are computed for each class separately in a multi-class classification problem, the accuracy A is computed for the entire test dataset usingand thus, out of all test samples, how many were correctly classified.
- Area Under ROC Curve (AUC): An ROC curve shows the performance of a classification model at different classification thresholds. The curve is computed by plotting the true positive rate () against the false positive rate () at these thresholds. The true positive rate for a bird species j is defined as:and the false positive rate for a bird species j is defined aswith denoting a probability threshold that is varied from 0 to 1 in order to obtain the ROC curve. The area under the ROC curve (AUC) gives an aggregate measure of the classification performance.The ROC was originally developed for a binary classifier and was later generalized for a multi-class classification system [36]. The test set labels are binarized by employing either the one-vs-one or the one-vs-rest configuration. We employed the one-vs-one configuration for our task. In more detail, different sound samples are ranked by their probabilities, and then false positive and true positive rates are computed by choosing different probability cut-offs to generate the ROC curve. The AUC is computed as the area under the ROC curve. In the end, an average across species is computed to get one AUC value for the entire data set, i.e.,
- Mean Average Precision (mAP): The evaluation metric gives us a way of characterizing the performance of a classifier by monitoring how precision changes with varying the classification probability threshold that the model uses to make a decision if a bird sound sample belongs to a class j. A good classifier will maintain a high precision as recall increases, while a poor classifier will take a hit on precision as recall increases with changes in threshold.In more detail, to compute Average Precision for a species j, a list of probabilities is generated in which the discrimination probabilities our model has assigned to all test samples for class j are stored. The list is then sorted by decreasing probabilities, and each element is assigned a rank k. By varying the rank k (by gradually lowering the probability threshold), a list of true positives and false positives is generated. Note that, as the classification threshold is lowered, the model labels increasingly more samples as positive. This will lead to an increase in false positives. The list is consequently employed to produce a list of precision values at different ranks . Considering all the K cases in the list where the sound sample belongs to class j, the average precision is computed as:where is an indicator function that equals unity if the sample at threshold k is a true positive. The mean average precision is then computed by averaging over all classes (species) [15].
3. Results and Discussion
3.1. Variations due to Randomized Sub-Sets
3.2. The Dependence on the Number of Species
3.3. Metric of Confidence
3.4. Comparing Different Measures for Classification Success
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| RMSE | root-mean-square energy |
| MFCC | Mel-frequency-cepstral-coefficients |
| ReLU | Rectified linear units |
| AUC | Area Under ROC Curve |
| ROC | Reciever Operating Characterics |
| mAP | Mean Average Precision |
| IQR | Interquartile range |
| ANN | Artificial neural netwroks |
References
- Sutherland, W.J.; Newton, I.; Green, R. Bird Ecology and Conservation: A Handbook of Techniques; OUP Oxford: Oxford, UK, 2004; Volume 1. [Google Scholar]
- Priyadarshani, N.; Marsland, S.; Castro, I. Automated birdsong recognition in complex acoustic environments: A review. J. Avian Biol. 2018, 49, jav-01447. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Li, Y. Adaptive energy detection for bird sound detection in complex environments. Neurocomputing 2015, 155, 108–116. [Google Scholar] [CrossRef]
- Jančovič, P.; Köküer, M. Automatic detection and recognition of tonal bird sounds in noisy environments. EURASIP J. Adv. Signal Process. 2011, 2011, 982936. [Google Scholar] [CrossRef] [Green Version]
- Fox, E.J.; Roberts, J.D.; Bennamoun, M. Text-independent speaker identification in birds. In Proceedings of the Ninth International Conference on Spoken Language Processing, Pittsburgh, PA, USA, 17–21 September 2006. [Google Scholar]
- Cai, J.; Ee, D.; Pham, B.; Roe, P.; Zhang, J. Sensor network for the monitoring of ecosystem: Bird species recognition. In Proceedings of the 2007 3rd International Conference on Intelligent Sensors, Sensor Networks and Information, Melbourne, VIC, Australia, 3–6 December 2007; pp. 293–298. [Google Scholar]
- Chen, Z.; Maher, R.C. Semi-automatic classification of bird vocalizations using spectral peak tracks. J. Acoust. Soc. Am. 2006, 120, 2974–2984. [Google Scholar] [CrossRef] [PubMed]
- Jančovič, P.; Köküer, M. Acoustic recognition of multiple bird species based on penalized maximum likelihood. IEEE Signal Process. Lett. 2015, 22, 1585–1589. [Google Scholar]
- Wielgat, R.; Potempa, T.; Świétojański, P.; Król, D. On using prefiltration in HMM-based bird species recognition. In Proceedings of the 2012 International Conference on Signals and Electronic Systems (ICSES), Wroclaw, Poland, 18–21 September 2012; pp. 1–5. [Google Scholar]
- Briggs, F.; Lakshminarayanan, B.; Neal, L.; Fern, X.Z.; Raich, R.; Hadley, S.J.; Hadley, A.S.; Betts, M.G. Acoustic classification of multiple simultaneous bird species: A multi-instance multi-label approach. J. Acoust. Soc. Am. 2012, 131, 4640–4650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juang, C.F.; Chen, T.M. Birdsong recognition using prediction-based recurrent neural fuzzy networks. Neurocomputing 2007, 71, 121–130. [Google Scholar] [CrossRef]
- Sprengel, E.; Jaggi, M.; Kilcher, Y.; Hofmann, T. Audio Based Bird Species Identification Using Deep Learning Techniques; Technical Report; CLEF: Evora, Portugal, 2016. [Google Scholar]
- Bastas, S.; Majid, M.W.; Mirzaei, G.; Ross, J.; Jamali, M.M.; Gorsevski, P.V.; Frizado, J.; Bingman, V.P. A novel feature extraction algorithm for classification of bird flight calls. In Proceedings of the 2012 IEEE International Symposium on Circuits and Systems (ISCAS), Seoul, Korea, 20–23 May 2012; pp. 1676–1679. [Google Scholar]
- Selin, A.; Turunen, J.; Tanttu, J.T. Wavelets in recognition of bird sounds. EURASIP J. Adv. Signal Process. 2006, 2007, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Kahl, S.; Stöter, F.R.; Goëau, H.; Glotin, H.; Planque, R.; Vellinga, W.P.; Joly, A. Overview of BirdCLEF 2019: Large-scale bird recognition in soundscapes. In Proceedings of the Working Notes of CLEF 2019-Conference and Labs of the Evaluation Forum, CEUR, Lugano, Switzerland, 9–12 September 2019; pp. 1–9. [Google Scholar]
- Xeno Canto. Available online: https://www.xeno-canto.org/ (accessed on 27 May 2020).
- McFee, B.; McVicar, M.; Balke, S.; Thomé, C.; Raffel, C.; Lee, D.; Nieto, O.; Battenberg, E.; Ellis, D.; Yamamoto, R.; et al. Librosa/Librosa: 0.6.3. 2019. Available online: https://doi.org/10.5281/zenodo (accessed on 21 June 2021).
- Marler, P.R.; Slabbekoorn, H. Nature’s Music: The Science of Birdsong; Elsevier: Milano, Italy, 2004. [Google Scholar]
- Kahl, S.; Wilhelm-Stein, T.; Klinck, H.; Kowerko, D.; Eibl, M. Recognizing birds from sound-the 2018 BirdCLEF baseline system. arXiv 2018, arXiv:1804.07177. [Google Scholar]
- Khalid, S.; Khalil, T.; Nasreen, S. A survey of feature selection and feature extraction techniques in machine learning. In Proceedings of the 2014 Science and Information Conference, London, UK, 27–29 August 2014; pp. 372–378. [Google Scholar]
- Xie, J.; Zhu, M. Handcrafted features and late fusion with deep learning for bird sound classification. Ecol. Informatics 2019, 52, 74–81. [Google Scholar] [CrossRef]
- Virtanen, T.; Plumbley, M.D.; Ellis, D. Computational Analysis of Sound Scenes and Events; Springer: Cham, Switzerland, 2018. [Google Scholar]
- Klapuri, A.; Davy, M. Signal Processing Methods for Music Transcription; Springer: New York, NY, USA, 2007. [Google Scholar]
- Smith, J.O. Spectral Audio Signal Processing, 2011 ed.; Online Book; W3K Publishing: Palo Alto, CA, USA, 2011; Available online: http://ccrma.stanford.edu/~jos/sasp/ (accessed on 23 July 2021).
- Giannakopoulos, T.; Pikrakis, A. Introduction to Audio Analysis: A MATLAB® Approach; Academic Press: Cambridge, MA, USA, 2014. [Google Scholar]
- Abreha, G.T. An Environmental Audio-Based Context Recognition System Using Smartphones. Master’s Thesis, University of Twente, Enschede, The Netherlands, 2014. [Google Scholar]
- Logan, B. Mel Frequency Cepstral Coefficients for Music Modeling; ISMIR: Plymouth, MA, USA, 2000; Volume 270, pp. 1–11. [Google Scholar]
- Abadi, M.; Barham, P.; Chen, J.; Chen, Z.; Davis, A.; Dean, J.; Devin, M.; Ghemawat, S.; Irving, G.; Isard, M.; et al. Tensorflow: A system for large-scale machine learning. In Proceedings of the 12th {USENIX} Symposium on Operating Systems Design and Implementation ({OSDI} 16), Savannah, GA, USA, 2–4 November 2016; pp. 265–283. [Google Scholar]
- Bebis, G.; Georgiopoulos, M. Feed-forward neural networks. IEEE Potentials 1994, 13, 27–31. [Google Scholar] [CrossRef]
- Fine, T.L. Feedforward Neural Network Methodology; Springer Science & Business Media: New York, NY, USA, 2006. [Google Scholar]
- Goodfellow, I.; Bengio, Y.; Courville, A.; Bengio, Y. Deep Learning; MIT Press: Cambridge, MA, USA, 2016; Volume 1. [Google Scholar]
- Bishop, C.M. Pattern Recognition and Machine Learning; Springer: Berlin/Heidelberg, Germany, 2006. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Performance metrics for classification problems in machine learning. Available online: https://medium.com/thalusai/performance-metrics-for-classification-problems-in-machine-learningpart-i-b085d432082b (accessed on 15 July 2021).
- Grandini, M.; Bagli, E.; Visani, G. Metrics for Multi-Class Classification: An Overview. arXiv 2020, arXiv:stat.ML/2008.05756. [Google Scholar]
- Hand, D.J.; Till, R.J. A simple generalisation of the area under the ROC curve for multiple class classification problems. Mach. Learn. 2001, 45, 171–186. [Google Scholar] [CrossRef]
- Nuzzo, R.L. The box plots alternative for visualizing quantitative data. PM&R 2016, 8, 268–272. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghani, B.; Hallerberg, S. A Randomized Bag-of-Birds Approach to Study Robustness of Automated Audio Based Bird Species Classification. Appl. Sci. 2021, 11, 9226. https://doi.org/10.3390/app11199226
Ghani B, Hallerberg S. A Randomized Bag-of-Birds Approach to Study Robustness of Automated Audio Based Bird Species Classification. Applied Sciences. 2021; 11(19):9226. https://doi.org/10.3390/app11199226
Chicago/Turabian StyleGhani, Burooj, and Sarah Hallerberg. 2021. "A Randomized Bag-of-Birds Approach to Study Robustness of Automated Audio Based Bird Species Classification" Applied Sciences 11, no. 19: 9226. https://doi.org/10.3390/app11199226





