Transcriptome Analyses of In Vitro Exercise Models by Clenbuterol Supplementation or Electrical Pulse Stimulation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Electrical Pulse Stimulation (EPS)
2.3. RNA Extraction
2.4. Quantitative Real-Time PCR Analysis (qRT-PCR)
2.5. RNA-Sequence
2.6. Statistical Analysis
3. Results
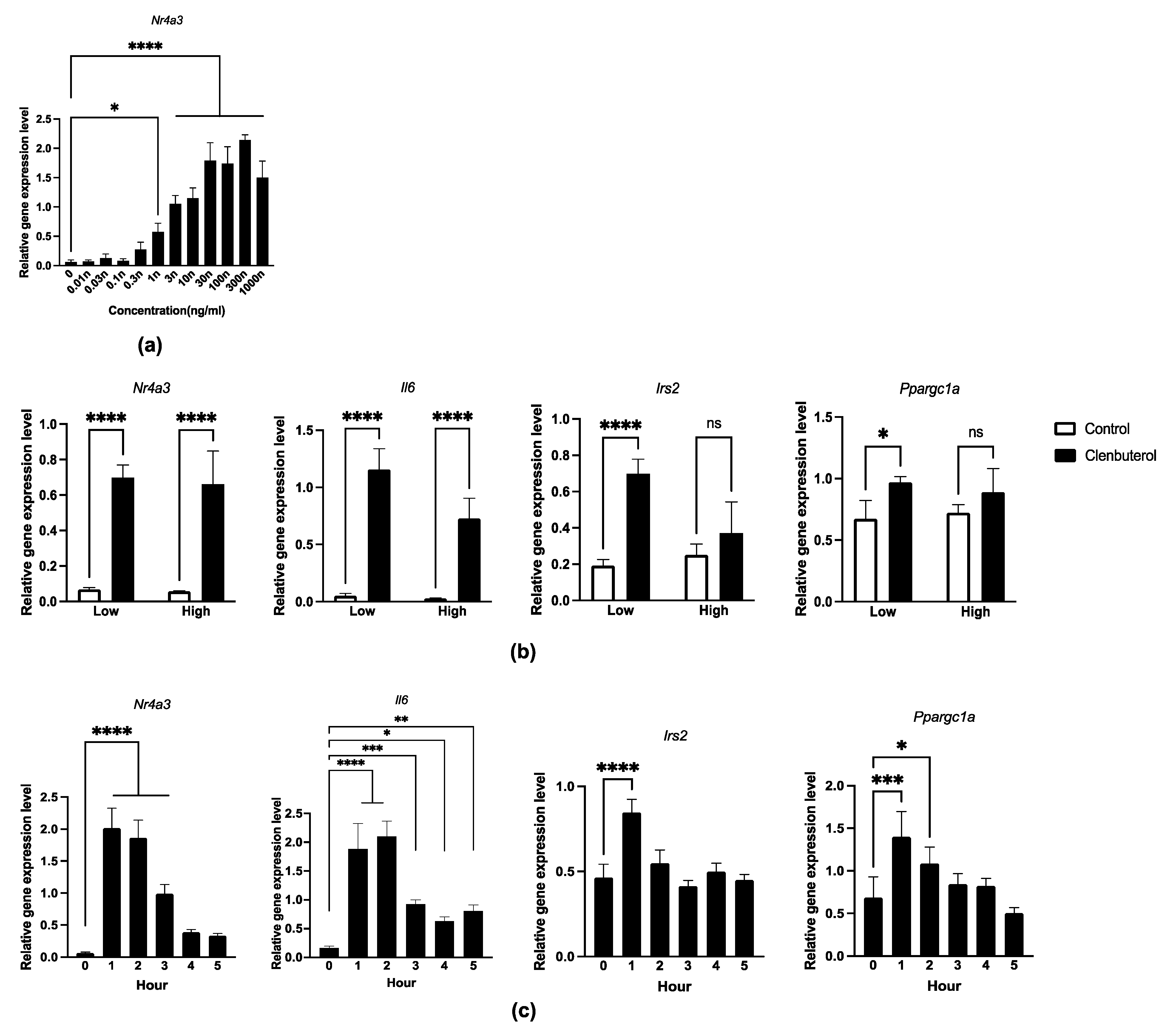
3.1. Acute Transcriptional Induction by Clenbuterol Supplementation
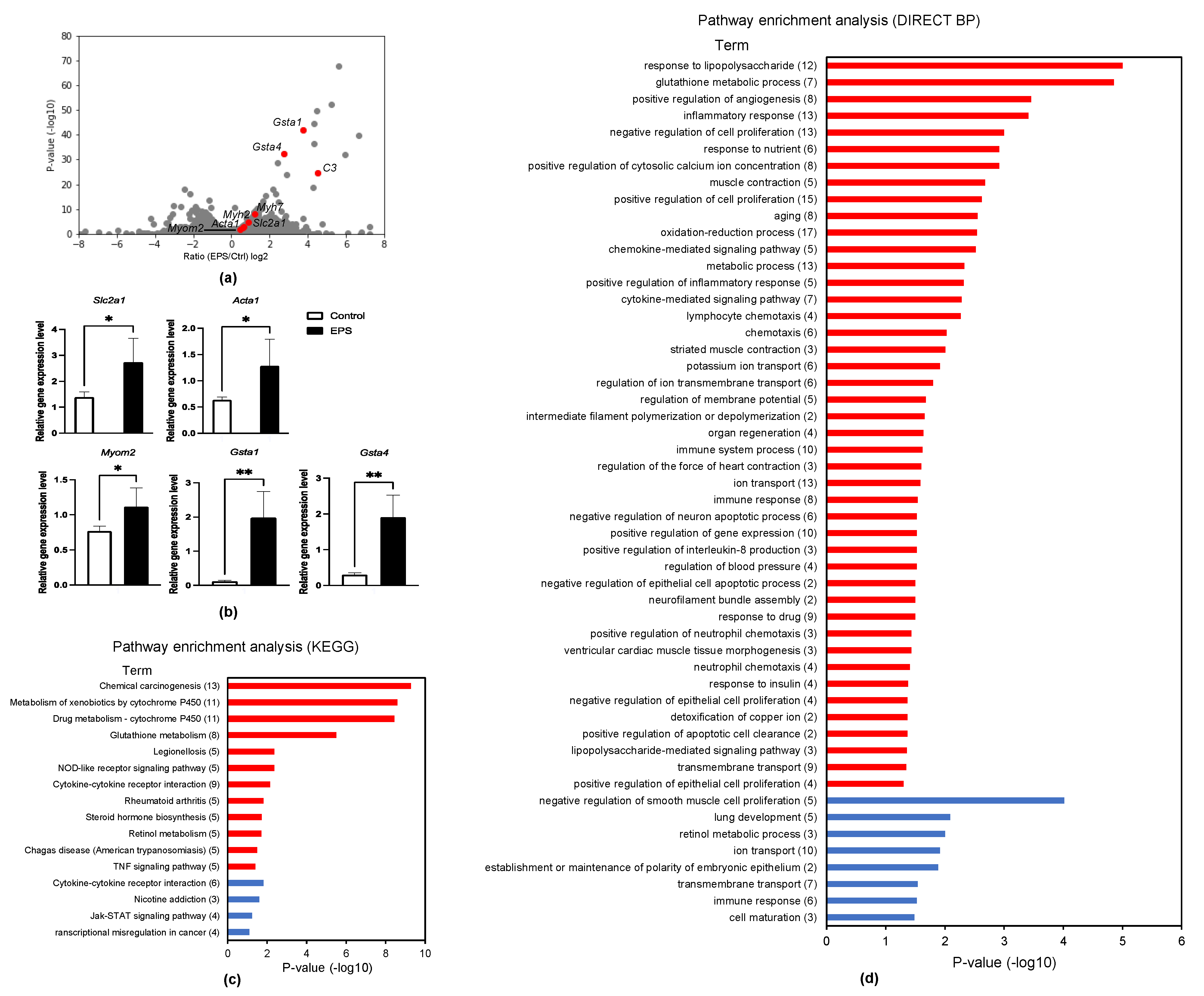
3.2. RNA-Seq Analysis in Clenbuterol-Supplemented Myotubes
3.3. Twenty-Four-Hour Electrical Pulse Stimulation in C2C12 Myotubes Induced Myh7 and Myh2
3.4. RNA-Seq Analysis in EPS Myotubes
3.5. IPA Analysis
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aldana, S.G.; Greenlaw, R.L.; Diehl, H.A.; Salberg, A.; Merrill, R.M.; Ohmine, S.; Thomas, C. Effects of an intensive diet and physical activity modification program on the health risks of adults. J. Am. Diet. Assoc. 2005, 105, 371–381. [Google Scholar] [CrossRef]
- Pedersen, B.K.; Saltin, B. Exercise as medicine—Evidence for prescribing exercise as therapy in 26 different chronic diseases. Scand. J. Med. Sci. Sport. 2015, 25, 1–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hegde, B.M. Health benefits of exercise. Kuwait Med. J. 2018, 50, 143–145. [Google Scholar] [CrossRef]
- Handschin, C.; Spiegelman, B.M. The role of exercise and PGC1α in inflammation and chronic disease. Nature 2008, 454, 463–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mancinelli, R.; Toniolo, L.; Di Filippo, E.S.; Doria, C.; Marrone, M.; Maroni, C.R.; Verratti, V.; Bondi, D.; Maccatrozzo, L.; Pietrangelo, T.; et al. Neuromuscular Electrical Stimulation Induces Skeletal Muscle Fiber Remodeling and Specific Gene Expression Profile in Healthy Elderly. Front. Physiol. 2019, 10, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Filippo, E.S.; Mancinelli, R.; Marrone, M.; Doria, C.; Verratti, V.; Toniolo, L.; Dantas, J.L.; Fulle, S.; Pietrangelo, T. Neuromuscular electrical stimulation improves skeletal muscle regeneration through satellite cell fusion with myofibers in healthy elderly subjects. J. Appl. Physiol. 2017, 123, 501–512. [Google Scholar] [CrossRef] [Green Version]
- Chapman, M.A.; Arif, M.; Emanuelsson, E.B.; Reitzner, S.M.; Lindholm, M.E.; Mardinoglu, A.; Sundberg, C.J. Skeletal Muscle Transcriptomic Comparison between Long-Term Trained and Untrained Men and Women. Cell Rep. 2020, 31. [Google Scholar] [CrossRef]
- Pillon, N.J.; Gabriel, B.M.; Dollet, L.; Smith, J.A.B.; Sardón Puig, L.; Botella, J.; Bishop, D.J.; Krook, A.; Zierath, J.R. Transcriptomic profiling of skeletal muscle adaptations to exercise and inactivity. Nat. Commun. 2020, 11. [Google Scholar] [CrossRef] [Green Version]
- Contrepois, K.; Wu, S.; Moneghetti, K.J.; Hornburg, D.; Ahadi, S.; Tsai, M.S.; Metwally, A.A.; Wei, E.; Lee-McMullen, B.; Quijada, J.V.; et al. Molecular Choreography of Acute Exercise. Cell 2020, 181, 1112–1130.e16. [Google Scholar] [CrossRef] [PubMed]
- Goto, M.; Terada, S.; Kato, M.; Katoh, M.; Yokozeki, T.; Tabata, I.; Shimokawa, T. cDNA Cloning and mRNA analysis of PGC-1 in epitrochlearis muscle in swimming-exercised rats. Biochem. Biophys. Res. Commun. 2000, 274, 350–354. [Google Scholar] [CrossRef]
- Booth, F.W.; Ruegsegger, G.N.; Toedebusch, R.G.; Yan, Z. Endurance Exercise and the Regulation of Skeletal Muscle Metabolism. Prog. Mol. Biol. Transl. Sci. 2015, 135, 129–151. [Google Scholar] [CrossRef]
- Schiaffino, S.; Reggiani, C. Fiber types in mammalian skeletal muscles. Physiol. Rev. 2011, 91, 1447–1531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- AHLQUIST, R.P. A study of the adrenotropic receptors. Am. J. Physiol. 1948, 153, 586–600. [Google Scholar] [CrossRef]
- Davis, E.; Loiacono, R.; Summers, R.J. The rush to adrenaline: Drugs in sport acting on the β-adrenergic system. Br. J. Pharmacol. 2008, 154, 584–597. [Google Scholar] [CrossRef] [Green Version]
- Awede, B.L.; Thissen, J.P.; Lebacq, J. Role of IGF-I and IGFBPs in the changes of mass and phenotype induced in rat soleus muscle by clenbuterol. Am. J. Physiol.-Endocrinol. Metab. 2002, 282, 31–37. [Google Scholar] [CrossRef] [Green Version]
- Abo, T.; Iida, R.H.; Kaneko, S.; Suga, T.; Yamada, H.; Hamada, Y.; Yamane, A. IGF and myostatin pathways are respectively induced during the earlier and the later stages of skeletal muscle hypertrophy induced by clenbuterol, a β2-adrenergic agonist. Cell Biochem. Funct. 2012, 30, 671–676. [Google Scholar] [CrossRef] [PubMed]
- Kitaoka, Y.; Watanabe, D.; Nonaka, Y.; Yagishita, K.; Kano, Y.; Hoshino, D. Effects of clenbuterol administration on mitochondrial morphology and its regulatory proteins in rat skeletal muscle. Physiol. Rep. 2019, 7, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miura, S.; Kawanaka, K.; Kai, Y.; Tamura, M.; Goto, M.; Shiuchi, T.; Minokoshi, Y.; Ezaki, O. An increase in murine skeletal muscle peroxisome proliferator-activated receptor-γ coactivator-1α (PGC-1α) mRNA in response to exercise is mediated by β-adrenergic receptor activation. Endocrinology 2007, 148, 3441–3448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikolić, N.; Görgens, S.W.; Thoresen, G.H.; Aas, V.; Eckel, J.; Eckardt, K. Electrical pulse stimulation of cultured skeletal muscle cells as a model for In Vitro exercise-possibilities and limitations. Acta Physiol. 2017, 220, 310–331. [Google Scholar] [CrossRef] [PubMed]
- Carter, S.; Solomon, T.P.J. In Vitro experimental models for examining the skeletal muscle cell biology of exercise: The possibilities, challenges and future developments. Pflugers Arch. 2019, 471, 413–429. [Google Scholar] [CrossRef] [Green Version]
- Hoshino, D.; Kawata, K.; Kunida, K.; Hatano, A.; Yugi, K.; Wada, T.; Fujii, M.; Sano, T.; Ito, Y.; Furuichi, Y.; et al. Trans-omic Analysis Reveals ROS-Dependent Pentose Phosphate Pathway Activation after High-Frequency Electrical Stimulation in C2C12 Myotubes. iScience 2020, 23, 101558. [Google Scholar] [CrossRef] [PubMed]
- Tamura, Y.; Kouzaki, K.; Kotani, T.; Nakazato, K.; Electrically, N.K. Making Cell Culture More Physiological Electrically stimulated contractile activity-induced transcriptomic responses and metabolic remodeling in C 2 C 12 myotubes: Twitch vs. tetanic contractions. Am. J. Physiol. Cell Physiol. 2020, 319, 1029–1044. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Pilegaard, H.; Osada, T.; Andersen, L.T.; Helge, J.W.; Saltin, B.; Neufer, P.D. Substrate availability and transcriptional regulation of metabolic genes in human skeletal muscle during recovery from exercise. Metabolism 2005, 54, 1048–1055. [Google Scholar] [CrossRef]
- Rundqvist, H.C.; Montelius, A.; Osterlund, T.; Norman, B.; Esbjornsson, M.; Jansson, E. Acute sprint exercise transcriptome in human skeletal muscle. PLoS ONE 2019, 14, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Myers, S.A.; Eriksson, N.; Burow, R.; Wang, S.C.M.; Muscat, G.E.O. β-Adrenergic signaling regulates NR4A nuclear receptor and metabolic gene expression in multiple tissues. Mol. Cell. Endocrinol. 2009, 309, 101–108. [Google Scholar] [CrossRef]
- Gordon, B.S.; Steiner, J.L.; Rossetti, M.L.; Qiao, S.; Ellisen, L.W.; Govindarajan, S.S.; Eroshkin, A.M.; Williamson, D.L.; Coen, P.M. REDD1 induction regulates the skeletal muscle gene expression signature following acute aerobic exercise. Am. J. Physiol. -Endocrinol. Metab. 2017, 313, E737–E747. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Fujita, H.; Nedachi, T.; Kanzaki, M. Accelerated de novo sarcomere assembly by electric pulse stimulation in C2C12 myotubes. Exp. Cell Res. 2007, 313, 1853–1865. [Google Scholar] [CrossRef]
- Nedachi, T.; Fujita, H.; Kanzaki, M. Contractile C2C12 myotube model for studying exercise-inducible responses in skeletal muscle. Am. J. Physiol.-Endocrinol. Metab. 2008, 295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burch, N.; Arnold, A.S.; Item, F.; Summermatter, S.; Santos, G.B.S.; Christe, M.; Boutellier, U.; Toigo, M.; Handschin, C. Electric pulse stimulation of cultured murine muscle cells reproduces gene expression changes of trained mouse muscle. PLoS ONE 2010, 5. [Google Scholar] [CrossRef] [Green Version]
- Meng, Z.X.; Gong, J.; Chen, Z.; Sun, J.; Xiao, Y.; Wang, L.; Li, Y.; Liu, J.; Xu, X.Z.S.; Lin, J.D. Glucose Sensing by Skeletal Myocytes Couples Nutrient Signaling to Systemic Homeostasis. Mol. Cell 2017, 66, 332–344.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheler, M.; Irmler, M.; Lehr, S.; Hartwig, S.; Staiger, H.; Al-Hasani, H.; Beckers, J.; de Angelis, M.H.; Häring, H.U.; Weigert, C. Cytokine response of primary human myotubes in an in vitro exercise model. Am. J. Physiol.-Cell Physiol. 2013, 305, 877–886. [Google Scholar] [CrossRef]
- Bintu, L.; Yong, J.; Antebi, Y.E.; McCue, K.; Kazuki, Y.; Uno, N.; Oshimura, M.; Elowitz, M.B. Dynamics of epigenetic regulation at the single-cell level. Science 2016, 351, 720–724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramachandran, K.; Senagolage, M.D.; Sommars, M.A.; Futtner, C.R.; Omura, Y.; Allred, A.L.; Barish, G.D. Dynamic Enhancers Control Skeletal Muscle Identity and Reprogramming. PLoS Biol. 2019, 17, e3000467. [Google Scholar] [CrossRef]
- Ohsawa, I.; Kawano, F. Chronic exercise training activates histone turnover in mouse skeletal muscle fibers. FASEB J. 2021, 35, 1–14. [Google Scholar] [CrossRef]
- Makhnovskii, P.A.; Zgoda, V.G.; Bokov, R.O.; Shagimardanova, E.I.; Gazizova, G.R.; Gusev, O.A.; Lysenko, E.A.; Kolpakov, F.A.; Vinogradova, O.L.; Popov, D.V. Regulation of Proteins in Human Skeletal Muscle: The Role of Transcription. Sci. Rep. 2020, 10, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popov, X.D.V.; Makhnovskii, P.A.; Shagimardanova, E.I.; Gazizova, G.R.; Lysenko, E.A.; Gusev, O.A.; Vinogradova, O.L. Contractile activity-specific transcriptome response to acute endurance exercise and training in human skeletal muscle. Am. J. Physiol.-Endocrinol. Metab. 2019, 316, E605–E614. [Google Scholar] [CrossRef]



| Gene Name | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| Actb | GGCTGTATTCCCCTCCATCG | CCAGTTGGTAACAATGCCATGT |
| Myh7 | AGGGCGACCTCAACGAGAT | CAGCAGACTCTGGAGGCTCTT |
| Myh2 | ACTTTGGCACTACGGGGAAAC | CAGCAGCATTTCGATCAGCTC |
| Myh4 | GCTTGAAAACGAGGTGGAAA | CCTCCTCAGCCTGTCTCTTG |
| Myh1 | TACTCACGCCAGCTAGACGA | TGCCTCTTCAGCTCCTCAAT |
| Irs2 | CTGCGTCCTCTCCCAAAGTG | GGGGTCATGGGCATGTAGC |
| Nr4a1 | TTGAGTTCGGCAAGCCTACC | GTGTACCCGTCCATGAAGGTG |
| Nr4a2 | GTGTTCAGGCGCAGTATGG | TGGCAGTAATTTCAGTGTTGGT |
| Nr4a3 | TCAGCCTTTTTGGAGCTGTT | TGAAGTCGATGCAGGACAAG |
| Ppargc1a | TATGGAGTGACATAGAGTGTGCT | CCACTTCAATCCACCCAGAAAG |
| Il6 | TAGTCCTTCCTACCCCAATTTCC | TTGGTCCTTAGCCACTCCTTC |
| Vegfa | TCTCTGGGAGAACCCAAATG | ATCCACCCACTAGGCAACAG |
| Cxcl1 | CTGGGATTCACCTCAAGAACATC | CAGGGTCAAGGCAAGCCTC |
| Cxcl5 | GTTCCATCTCGCCATTCATGC | GCGGCTATGACTGAGGAAGG |
| Per1 | CGGATTGTCTATATTTCGGAGCA | TGGGCAGTCGAGATGGTGTA |
| Slc2a1 | CAGTTCGGCTATAACACTGGTG | GCCCCCGACAGAGAAGATG |
| Acta1 | CCCAAAGCTAACCGGGAGAAG | CCAGAATCCAACACGATGCC |
| Gsta1 | AAGCCCGTGCTTCACTACTTC | GGGCACTTGGTCAAACATCAAA |
| Gsta4 | TGATTGCCGTGGCTCCATTTA | CAACGAGAAAAGCCTCTCCGT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fukushima, T.; Takata, M.; Kato, A.; Uchida, T.; Nikawa, T.; Sakakibara, I. Transcriptome Analyses of In Vitro Exercise Models by Clenbuterol Supplementation or Electrical Pulse Stimulation. Appl. Sci. 2021, 11, 10436. https://doi.org/10.3390/app112110436
Fukushima T, Takata M, Kato A, Uchida T, Nikawa T, Sakakibara I. Transcriptome Analyses of In Vitro Exercise Models by Clenbuterol Supplementation or Electrical Pulse Stimulation. Applied Sciences. 2021; 11(21):10436. https://doi.org/10.3390/app112110436
Chicago/Turabian StyleFukushima, Taku, Miho Takata, Ayano Kato, Takayuki Uchida, Takeshi Nikawa, and Iori Sakakibara. 2021. "Transcriptome Analyses of In Vitro Exercise Models by Clenbuterol Supplementation or Electrical Pulse Stimulation" Applied Sciences 11, no. 21: 10436. https://doi.org/10.3390/app112110436
APA StyleFukushima, T., Takata, M., Kato, A., Uchida, T., Nikawa, T., & Sakakibara, I. (2021). Transcriptome Analyses of In Vitro Exercise Models by Clenbuterol Supplementation or Electrical Pulse Stimulation. Applied Sciences, 11(21), 10436. https://doi.org/10.3390/app112110436







