Self-Assembled MoS2/ssDNA Nanostructures for the Capacitive Aptasensing of Acetamiprid Insecticide
Abstract
:Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Nanomaterial Preparation
2.2.1. Preparation of MoS2 Nanosheets
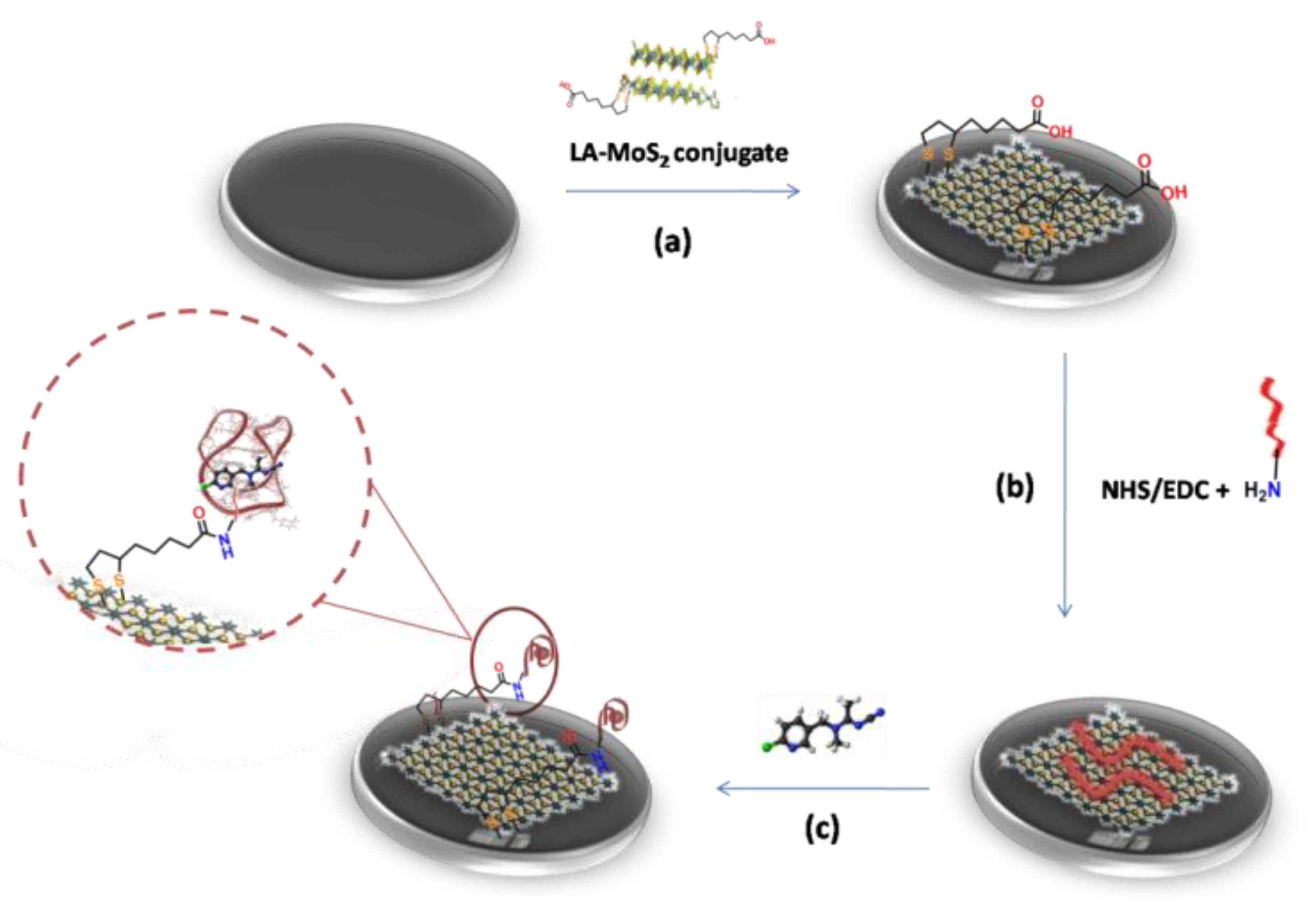
2.2.2. Preparation of LA-MoS2 Conjugate
2.3. Modification of SPCE with MoS2
2.4. Formation of Acetamiprid Aptasensor
2.5. Aptamer Binding to Acetamiprid
2.6. Electrochemical Measurements
2.7. Methods
3. Results and Discussions
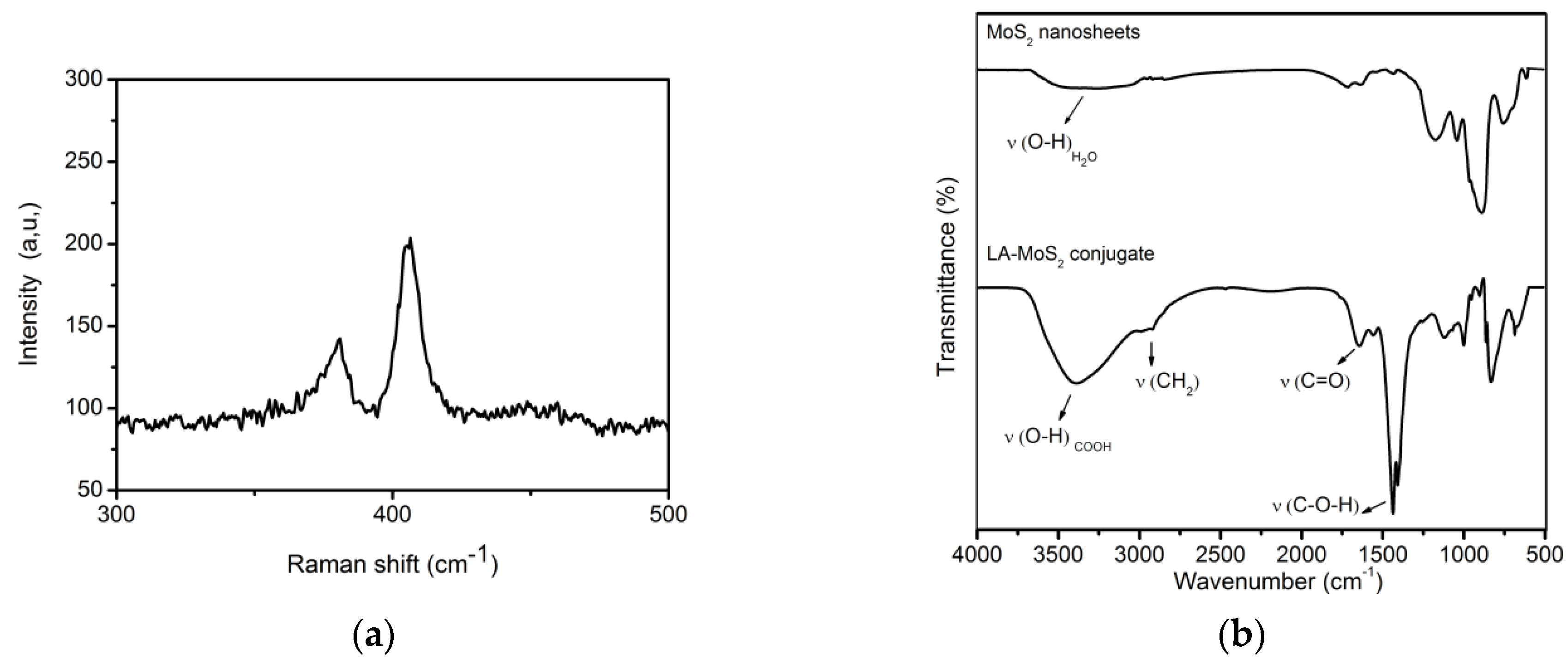
3.1. LA-MoS2 Synthesis and Structural Characterization
3.2. LA-MoS2/SPCE Surface Modification and Characterization
3.3. Biolayer Formation
3.4. Analytical Performances
3.4.1. Acetamiprid Detection
3.4.2. Comparison with Reported Acetamiprid Aptasensors
3.4.3. Selectivity
3.5. Detection of Acetamiprid in Fortified Tomatoe Sample
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Imamura, T.; Yanagawa, Y.; Nishikawa, K.; Matsumoto, N.; Sakamoto, T. Two cases of acute poisoning with acetamiprid in humans. Clin. Toxicol. 2010, 48, 851–853. [Google Scholar] [CrossRef] [PubMed]
- Kocaman, A.Y.; Topaktaş, M. Genotoxic effects of a particular mixture of acetamiprid and alpha-cypermethrin on chromosome aberration, sister chromatid exchange, and micronucleus formation in human peripheral blood lymphocytes. Environ. Toxicol. 2010, 25, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Xiu, F.-R.; Zheng, M.; Li, B. A simple and rapid chemiluminescence aptasensor for acetamiprid in contaminated samples: Sensitivity, selectivity and mechanism. Biosens. Bioelectron. 2016, 83, 243–249. [Google Scholar] [CrossRef] [PubMed]
- U.S. Environmental Protection Agency. Name of Chemical: Acetamiprid Reason for Issuance: Conditional Registration. 2002 Date Issued: March 15 Acetamiprid Proposed Interim Registration Review Decision Case Number 7617 January 2020, Docket Number EPA-HQ-OPP-2012-0329. Available online: https://www.epa.gov/sites/production/files/2020-01/documents/acetamiprid_pid.pdf (accessed on 9 September 2020).
- Wanatabe, S.; Ito, S.; Kamata, Y.; Omoda, N.; Yamazaki, T.; Munakata, H.; Kaneko, T.; Yuasa, Y. Development of competitive enzyme-linked immunosorbent assays (ELISAs) based on monoclonal antibodies for chloronicotinoid insecticides imidacloprid and acetamiprid. Anal. Chim. Acta 2001, 427, 211–219. [Google Scholar] [CrossRef]
- Zhang, B.; Pan, X.; Venne, L.; Dunnum, S.; McMurry, S.T.; Cobb, G.P.; Anderson, T.A. Development of a method for the determination of 9 currently used cotton pesticides by gas chromatography with electron capture detection. Talanta 2008, 75, 1055–1060. [Google Scholar] [CrossRef]
- Vichapong, J.; Burakham, R.; Srijaranai, S. Alternative Liquid–Liquid Microextraction as Cleanup for Determination of Neonicotinoid Pesticides Prior HPLC Analysis. Chromatographia 2016. [Google Scholar] [CrossRef]
- Mateu-Sánchez, M.; Moreno, M.; Arrebola, F.J.; Vidal, J.L.M. Analysis of Acetamiprid in Vegetables Using Gas Chromatography-Tandem Mass Spectrometry. Anal. Sci. 2003, 19, 701–704. [Google Scholar] [CrossRef] [Green Version]
- Zeng, L.; Peng, L.; Wu, D.; Yang, B. Electrochemical Sensors for Food Safety. In Nutrition in Health and Disease—Our Challenges Now and Forthcoming Time; IntechOpen: London, UK, 2018. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Mohamed, M.A.; Vinu Mohan, A.M.; Zhu, Z.; Sharma, V.; Mishra, G.K.; Mishra, R.K. Application of Electrochemical Aptasensors toward Clinical Diagnostics, Food, and Environmental Monitoring: Review. Sensors (Basel) 2019, 19, 5435. [Google Scholar] [CrossRef] [Green Version]
- Ben Aissa, S.; Mars, A.; Catanante, G.; Marty, J.-L.; Raouafi, N. Design of a redox-active surface for ultrasensitive redox capacitive aptasensing of aflatoxin M1 in milk. Talanta 2019, 195, 525–532. [Google Scholar] [CrossRef]
- Zou, X.; Wu, J.; Gu, J.; Shen, L.; Mao, L. Application of Aptamers in Virus Detection and Antiviral Therapy. Front. Microbiol. 2019, 10, 1462. [Google Scholar] [CrossRef] [Green Version]
- Raouafi, A.; Sánchez, A.; Raouafi, N.; Villalonga, R. Electrochemical aptamer-based bioplatform for ultrasensitive detection of prostate specific antigen. Sens. Actuators B Chem. 2019, 297, 126762. [Google Scholar] [CrossRef]
- Sun, D.; Lu, J.; Zhang, L.; Chen, Z. Aptamer-based electrochemical cytosensors for tumor cell detection in cancer diagnosis: A review. Anal. Chim. Acta 2019, 1082, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Sun, J.; Yao, Y.; Liu, H.; Huang, J.; Guo, Y.; Sun, X. Novel electrochemical aptasensor with dual signal amplification strategy for detection of acetamiprid. Sci. Total Environ. 2020, 705, 135905. [Google Scholar] [CrossRef] [PubMed]
- Taghdisi, S.M.; Danesh, N.M.; Ramezani, M.; Abnous, K. Electrochemical aptamer based assay for the neonicotinoid insecticide acetamiprid based on the use of an unmodified gold electrode. Microchim. Acta 2017, 184, 499–505. [Google Scholar] [CrossRef]
- Fei, A.; Liu, Q.; Huan, J.; Qian, J.; Dong, X.; Qiu, B.; Mao, H.; Wang, K. Label-free impedimetric aptasensor for detection of femtomole level acetamiprid using gold nanoparticles decorated multiwalled carbon nanotube-reduced graphene oxide nanoribbon composites. Biosens. Bioelectron. 2015, 70, 122–129. [Google Scholar] [CrossRef]
- Santos, A. Fundamentals and Applications of Impedimetric and Redox Capacitive Biosensors. J. Anal. Bioanal. Tech. 2014, S7. [Google Scholar] [CrossRef]
- Cecchetto, J.; Fernandes, F.C.B.; Lopes, R.; Bueno, P.R. The capacitive sensing of NS1 Flavivirus biomarker. Biosens. Bioelectron. 2017, 87, 949–956. [Google Scholar] [CrossRef] [Green Version]
- Lehr, J.; Weeks, J.R.; Santos, A.; Feliciano, G.T.; Nicholson, M.I.G.; Davis, J.J.; Bueno, P.R. Mapping the ionic fingerprints of molecular monolayers. Phys. Chem. Chem. Phys. 2017, 19, 15098–15109. [Google Scholar] [CrossRef]
- Raouafi, A.; Rabti, A.; Raouafi, N. A printed SWCNT electrode modified with polycatechol and lysozyme for capacitive detection of α-lactalbumin. Microchim. Acta 2017, 184, 4351–4357. [Google Scholar] [CrossRef]
- Fernandes, F.C.B.; Góes, M.S.; Davis, J.J.; Bueno, P.R. Label free redox capacitive biosensing. Biosens. Bioelectron. 2013, 50, 437–440. [Google Scholar] [CrossRef]
- Su, S.; Sun, Q.; Gu, X.; Xu, Y.; Shen, J.; Zhu, D.; Chao, J.; Fan, C.; Wang, L. Two-dimensional nanomaterials for biosensing applications. TrAC Trends Anal. Chem. 2019, 119, 115610. [Google Scholar] [CrossRef]
- Patel, R.; Inamdar, A.; Kim, H.; Im, H. In-Situ hydrothermal synthesis of a MoS2 nanosheet electrode for electrochemical energy storage applications. J. Korean Phys. Soc. 2016, 68, 1341–1346. [Google Scholar] [CrossRef]
- Soon, J.M.; Loh, K. Electrochemical Double-Layer Capacitance of MoS2 Nanowall Films. Electrochem. Solid-State Lett. 2007, 10, A250. [Google Scholar] [CrossRef]
- Chen, X.; Park, Y.J.; Kang, M.; Kang, S.-K.; Koo, J.; Shinde, S.M.; Shin, J.; Jeon, S.; Park, G.; Yan, Y.; et al. CVD-grown monolayer MoS 2 in bioabsorbable electronics and biosensors. Nat. Commun. 2018, 9, 1690. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Chen, X.; Wang, Q.; Xiao, M.; Zhong, D.; Sun, W.; Zhang, G.; Zhang, Z. Ultrasensitive Monolayer MoS2 Field-Effect Transistor Based DNA Sensors for Screening of Down Syndrome. Nano Lett. 2019, 19, 1437–1444. [Google Scholar] [CrossRef] [PubMed]
- Dalila R, N.; Md Arshad, M.K.; Gopinath, S.C.B.; Norhaimi, W.M.W.; Fathil, M.F.M. Current and future envision on developing biosensors aided by 2D molybdenum disulfide (MoS2) productions. Biosens. Bioelectron. 2019, 132, 248–264. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Zeng, Z.; Li, H.; Li, F.; Fan, C.; Zhang, H. Single-Layer MoS2-Based Nanoprobes for Homogeneous Detection of Biomolecules. J. Am. Chem. Soc. 2013, 135, 5998–6001. [Google Scholar] [CrossRef]
- He, J.; Liu, Y.; Fan, M.; Liu, X. Isolation and Identification of the DNA Aptamer Target to Acetamiprid. J. Agric. Food Chem. 2011, 59, 1582–1586. [Google Scholar] [CrossRef]
- Zhang, X.H.; Wang, C.; Xue, M.Q.; Lin, B.C.; Ye, X.; Lei, W.N. Hydrothermal synthesis and characterization of ultrathin MoS2 nanosheets. Chalcogenide Lett. 2016, 13, 27–34. [Google Scholar] [CrossRef] [Green Version]
- Jeong, M.; Kim, S.; Ju, S.-Y. Preparation and characterization of a covalent edge-functionalized lipoic acid–MoS2 conjugate. RSC Adv. 2016, 6, 36248–36255. [Google Scholar] [CrossRef]
- Haddaoui, M.; Sola, C.; Raouafi, N.; Korri-Youssoufi, H. E-DNA detection of rpoB gene resistance in Mycobacterium tuberculosis in real samples using Fe3O4/polypyrrole nanocomposite. Biosens. Bioelectron. 2019, 128, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Feier, B.; Băjan, I.; Cristea, C.; Săndulescu, R. Aptamer-based Electrochemical Sensor for the Detection of Ampicillin. In Proceedings of the International Conference on Advancements of Medicine and Health Care through Technology, Cluj-Napoca, Romania, 12–15 October 2016; Vlad, S., Roman, N.M., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 107–110. [Google Scholar]
- Li, H.; Lu, G.; Yin, Z.; He, Q.; Li, H.; Zhang, Q.; Zhang, H. Optical Identification of Single- and Few-Layer MoS2 Sheets. Small 2012, 8, 682–686. [Google Scholar] [CrossRef] [PubMed]
- Nie, C.; Zhang, B.; Gao, Y.; Yin, M.; Yi, X.; Zhao, C.; Zhang, Y.; Luo, L.; Wang, S. Thickness-Dependent Enhancement of Electronic Mobility of MoS2 Transistors via Surface Functionalization. J. Phys. Chem. C 2020, 124, 16943–16950. [Google Scholar] [CrossRef]
- Zhou, K.; Jiang, S.; Bao, C.; Song, L.; Wang, B.; Tang, G.; Hu, Y.; Gui, Z. Preparation of poly(vinyl alcohol) nanocomposites with molybdenum disulfide (MoS2): Structural characteristics and markedly enhanced properties. RSC Adv. 2012, 2, 11695. [Google Scholar] [CrossRef]
- Kong, N.; Zhang, S.; Liu, J.; Wang, J.; Liu, Z.; Wang, H.; Liu, J.; Yang, W. The influence of 2D nanomaterials on electron transfer across molecular thin films. Mol. Syst. Des. Eng. 2019, 4, 431–436. [Google Scholar] [CrossRef]
- Rastogi, P.K.; Sarkar, S.; Mandler, D. Ionic strength induced electrodeposition of two-dimensional layered MoS2 nanosheets. Appl. Mater. Today 2017, 8, 44–53. [Google Scholar] [CrossRef]
- Mejri-Omrani, N.; Miodek, A.; Zribi, B.; Marrakchi, M.; Hamdi, M.; Marty, J.-L.; Korri-Youssoufi, H. Direct detection of OTA by impedimetric aptasensor based on modified polypyrrole-dendrimers. Anal. Chim. Acta 2016, 920, 37–46. [Google Scholar] [CrossRef]
- Wang, Y.; Ning, G.; Bi, H.; Wu, Y.; Liu, G.; Zhao, Y. A Novel Ratiometric Electrochemical Assay for Ochratoxin A Coupling Au Nanoparticles Decorated MoS2 Nanosheets with Aptamer. Electrochim. Acta 2018, 285, 120–127. [Google Scholar] [CrossRef]
- Yao, Y.; Wang, G.X.; Shi, X.J.; Li, J.S.; Yang, F.Z.; Cheng, S.T.; Zhang, H.; Dong, H.W.; Guo, Y.M.; Sun, X.; et al. Ultrasensitive aptamer-based biosensor for acetamiprid using tetrahedral DNA nanostructures. J. Mater. Sci. 2020, 55, 15975–15987. [Google Scholar] [CrossRef]
- Madianos, L.; Tsekenis, G.; Skotadis, E.; Patsiouras, L.; Tsoukalas, D. A highly sensitive impedimetric aptasensor for the selective detection of acetamiprid and atrazine based on microwires formed by platinum nanoparticles. Biosens. Bioelectron. 2018, 101, 268–274. [Google Scholar] [CrossRef]
- Jiang, D.; Du, X.; Liu, Q.; Zhou, L.; Dai, L.; Qian, J.; Wang, K. Silver nanoparticles anchored on nitrogen-doped graphene as a novel electrochemical biosensing platform with enhanced sensitivity for aptamer-based pesticide assay. Analyst 2015, 140, 6404–6411. [Google Scholar] [CrossRef] [PubMed]
- Grimalt, S.; Dehouck, P. Review of Analytical Methods for the Determination of Pesticide Residues in Grapes. J. Chromatogr. A 2016, 1433, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-J.; Shelver, W.L.; Li, Q.X. Monoclonal antibody-based enzyme-linked immunosorbent assay for the insecticide imidacloprid. Anal. Chim. Acta 2004, 509, 111–118. [Google Scholar] [CrossRef]






| Platforms | Detection Method | Dynamic Range | LOD | Ref |
|---|---|---|---|---|
| GCE 1/AuNPs 2 | CV | 0.1 pM–10 nM | 0.077 pM | [42] |
| PtNPs 3 microstrips modified Au IDEs 4 | EIS | 10 pM–100 nM | 1 pM | [43] |
| SiNP 5-streptavidin conjugate modified MB-dsDNA 6 | DPV | 500 pM–6.5 µM | 1.53 pM | [16] |
| GCE/rGO-AgNPs 7/PB-AuNPs 8 | CV | 1 pM–1 µM | 0.3 pM | [15] |
| GCE/Au/MWCNT-rGONRe 9 | EIS | 50 fM–10 µM | 17 fM | [17] |
| GCE/Ag NPs anchored on nitrogen-doped graphene | EIS | 100 fM–5 nM | 33 fM | [44] |
| SPCE/LA-MoS2 | ECS | 50 fM–450 fM | 14 fM | This work |
| Sample | [ACE]added | [ACE]found | Recovery (%) |
|---|---|---|---|
| 1 ci | 65.0 | 62.1 ± 0.51 | 95.5 |
| 2 (ci + 55.0) | 120.0 | 120.2 ± 0.28 | 100.1 |
| 3 (ci + 155.0) | 220.0 | 227.0 ± 0.32 | 103.2 |
| 4 (ci + 255.0) | 320.0 | 334.0 ± 0.45 | 104.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hamami, M.; Raouafi, N.; Korri-Youssoufi, H. Self-Assembled MoS2/ssDNA Nanostructures for the Capacitive Aptasensing of Acetamiprid Insecticide. Appl. Sci. 2021, 11, 1382. https://doi.org/10.3390/app11041382
Hamami M, Raouafi N, Korri-Youssoufi H. Self-Assembled MoS2/ssDNA Nanostructures for the Capacitive Aptasensing of Acetamiprid Insecticide. Applied Sciences. 2021; 11(4):1382. https://doi.org/10.3390/app11041382
Chicago/Turabian StyleHamami, Maroua, Noureddine Raouafi, and Hafsa Korri-Youssoufi. 2021. "Self-Assembled MoS2/ssDNA Nanostructures for the Capacitive Aptasensing of Acetamiprid Insecticide" Applied Sciences 11, no. 4: 1382. https://doi.org/10.3390/app11041382






