Non-Invasive Measurement, Mathematical Simulation and In Situ Detection of Biofilm Evolution in Porous Media: A Review
Abstract
:1. Introduction
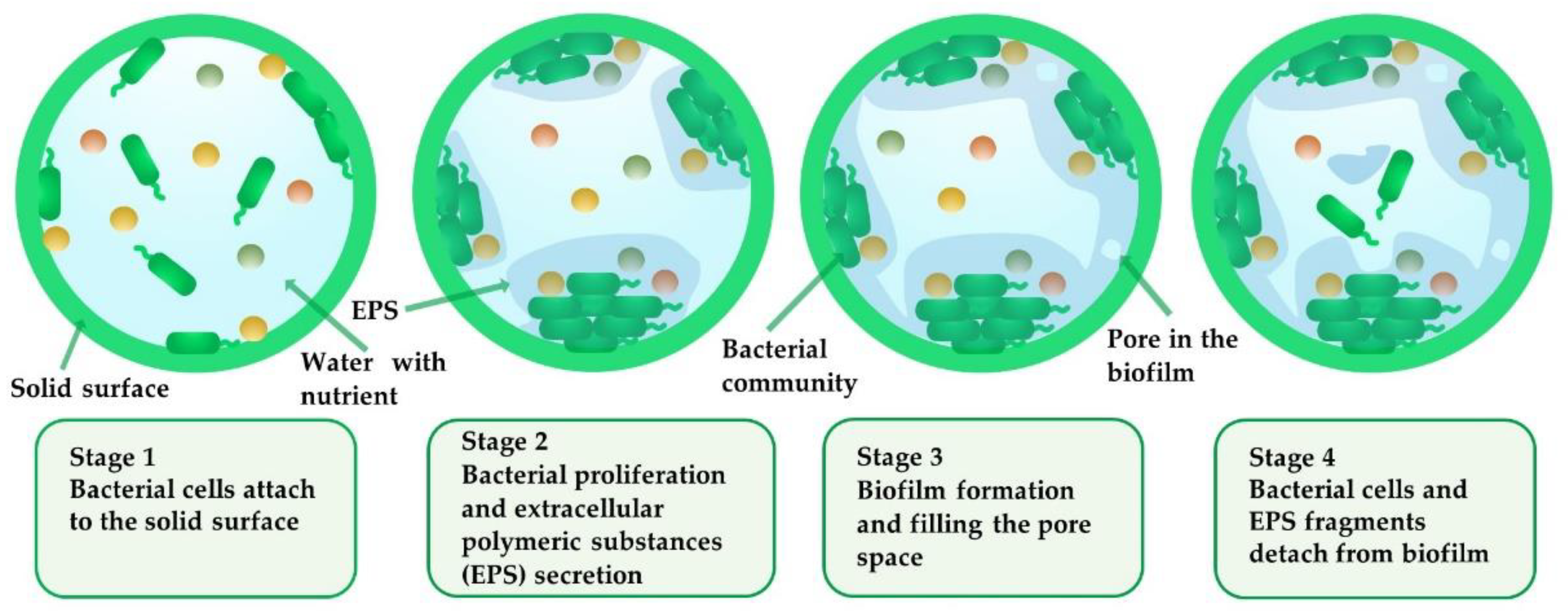
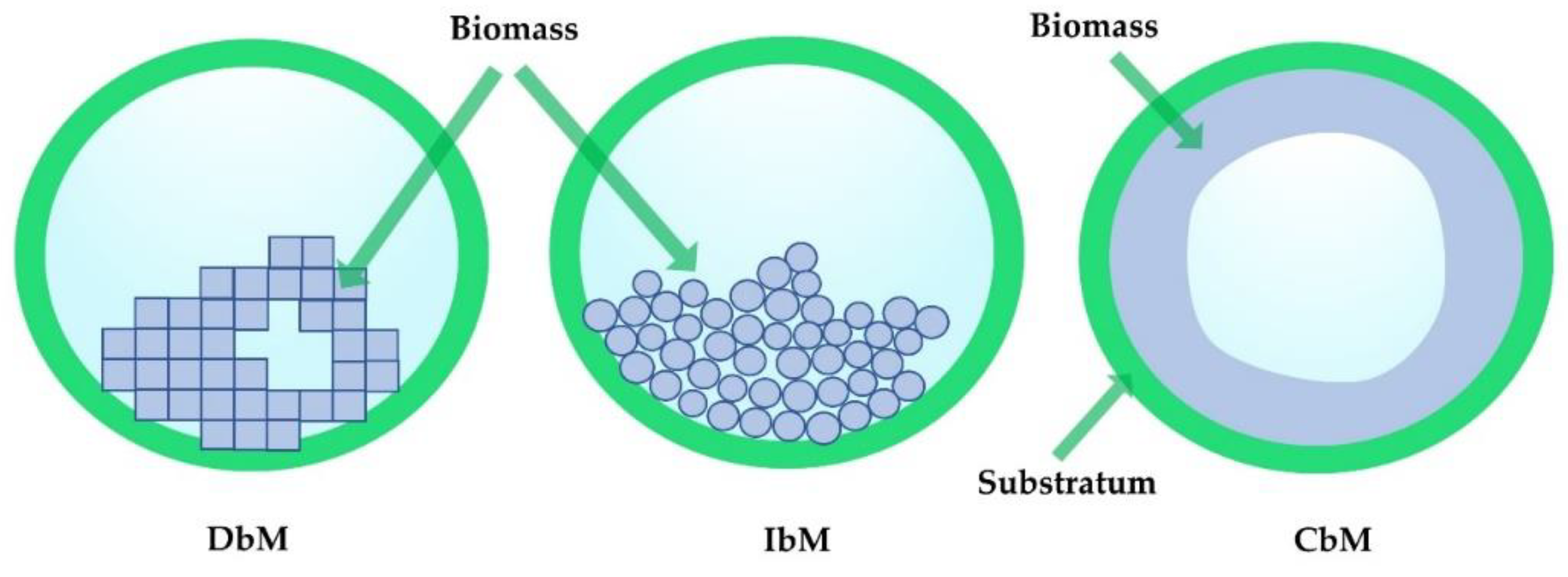
2. Characteristics of Biofilm Growth in Porous Media
3. Non-Invasive and In Situ Measurement of Mass Transport and Fluid Hydrodynamics of Bioclogging System in Porous Media
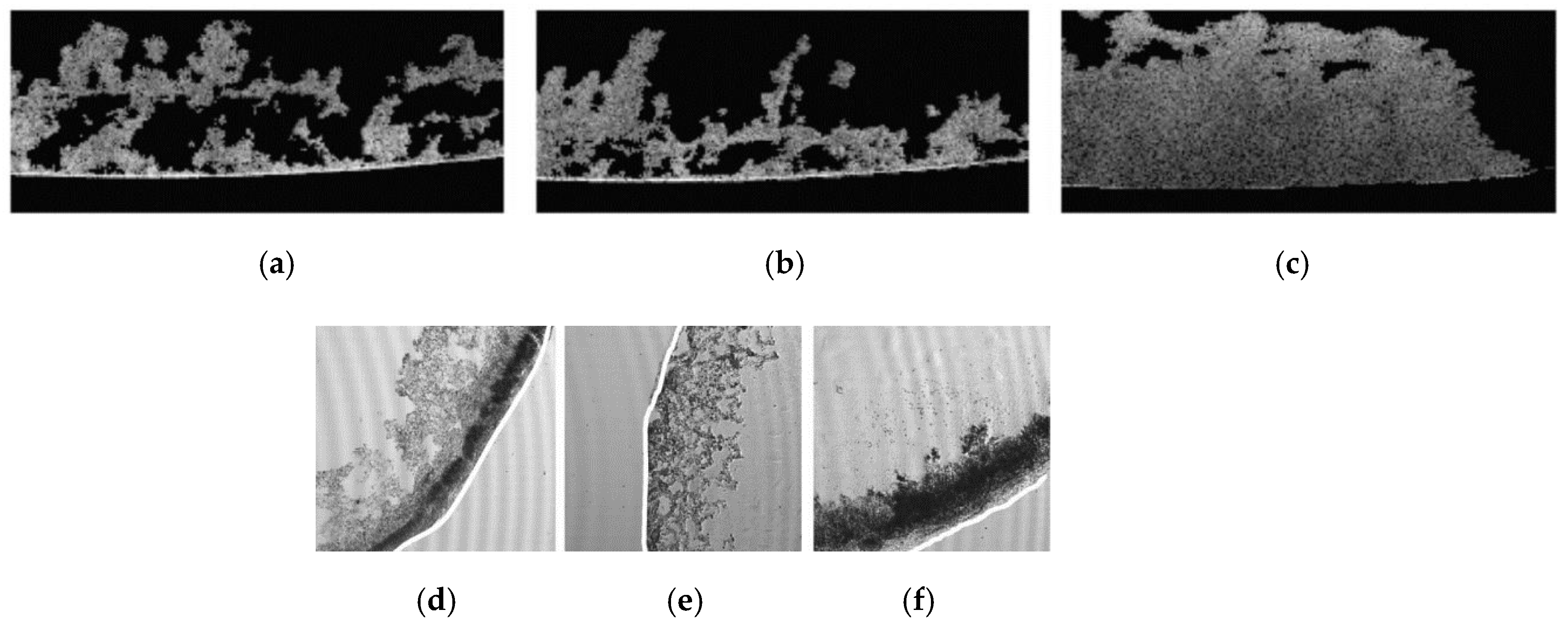
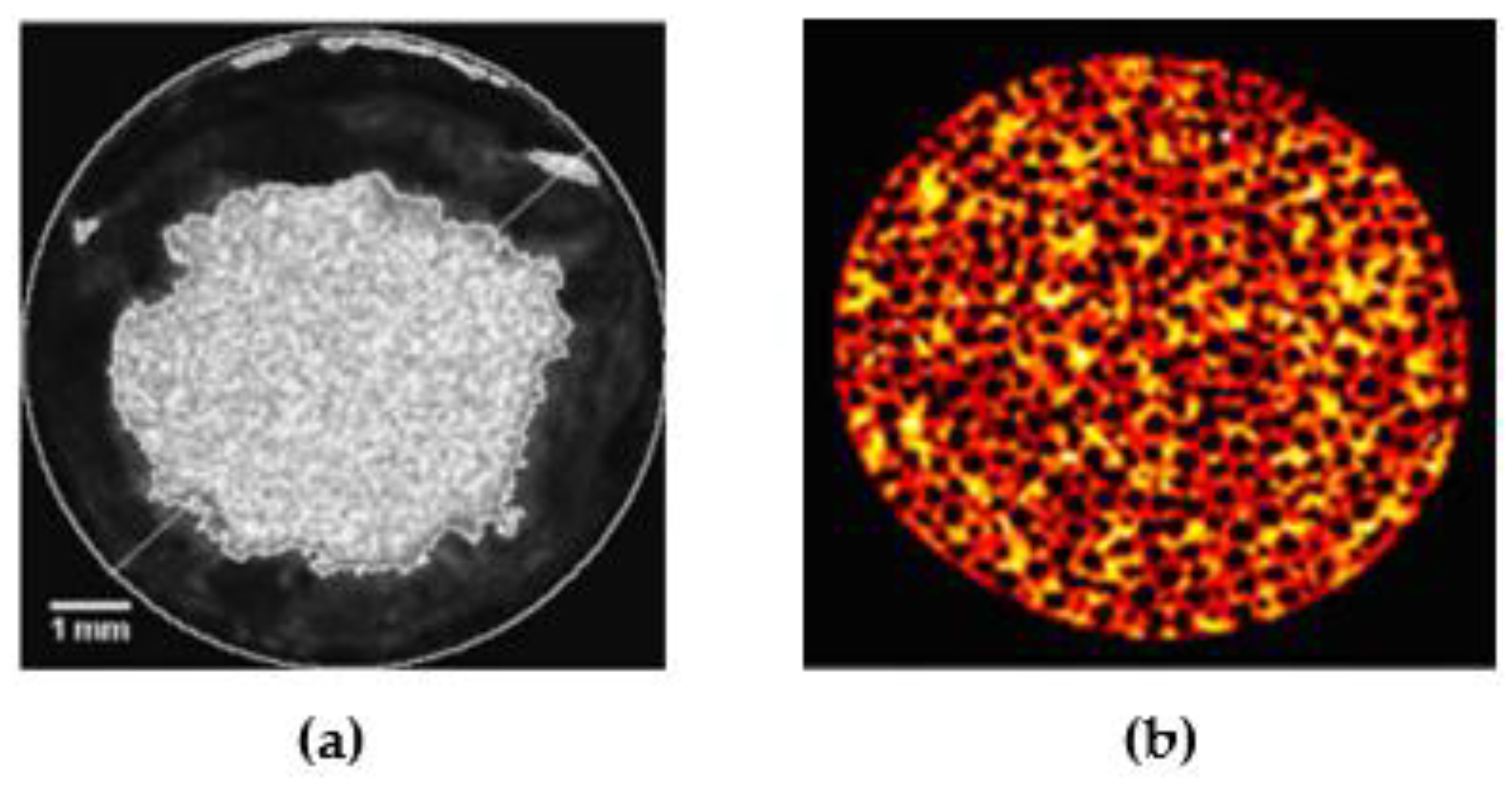
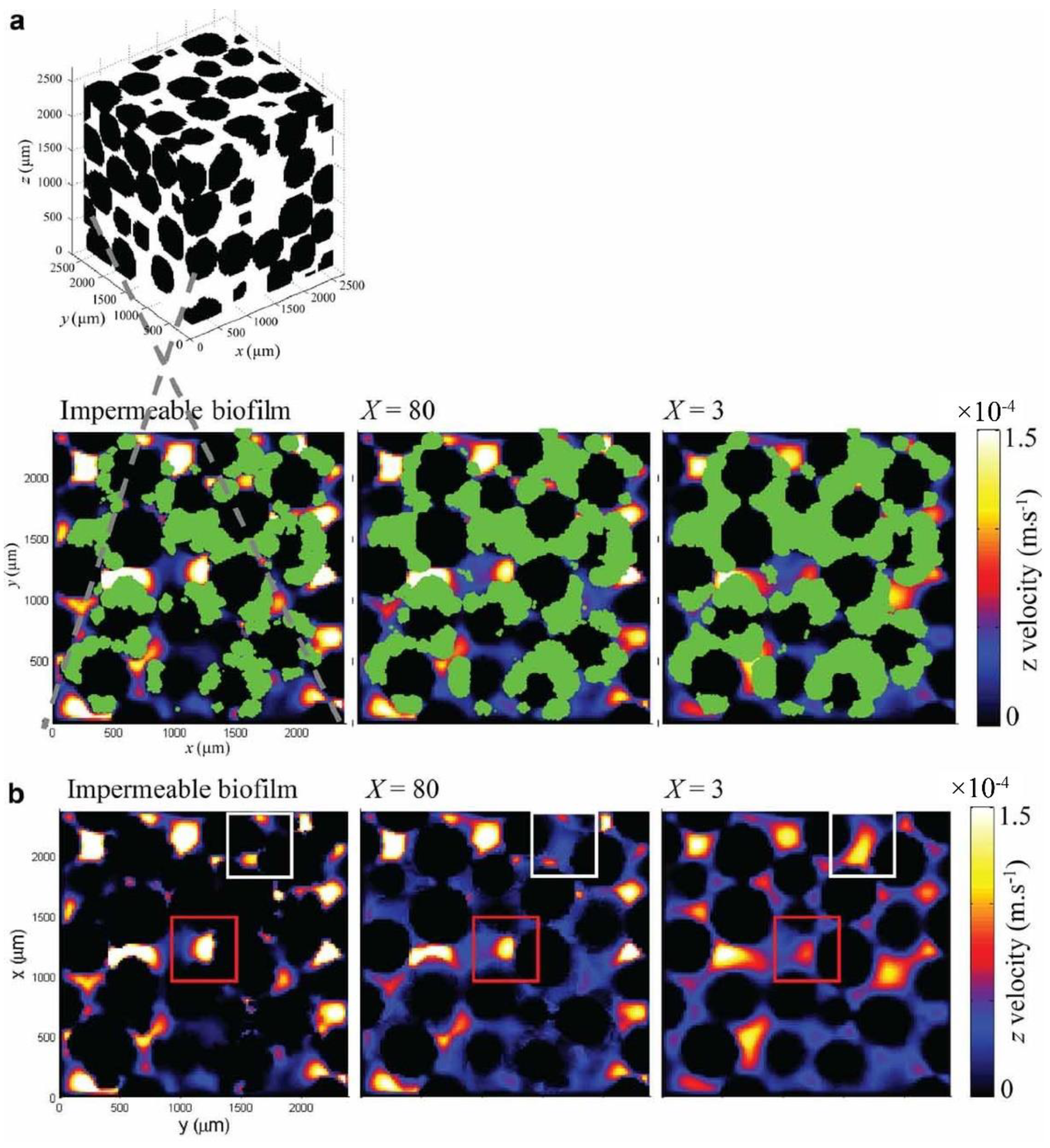
3.1. X-ray Tomography Technology
3.2. Nuclear Magnetic Resonance Measurement
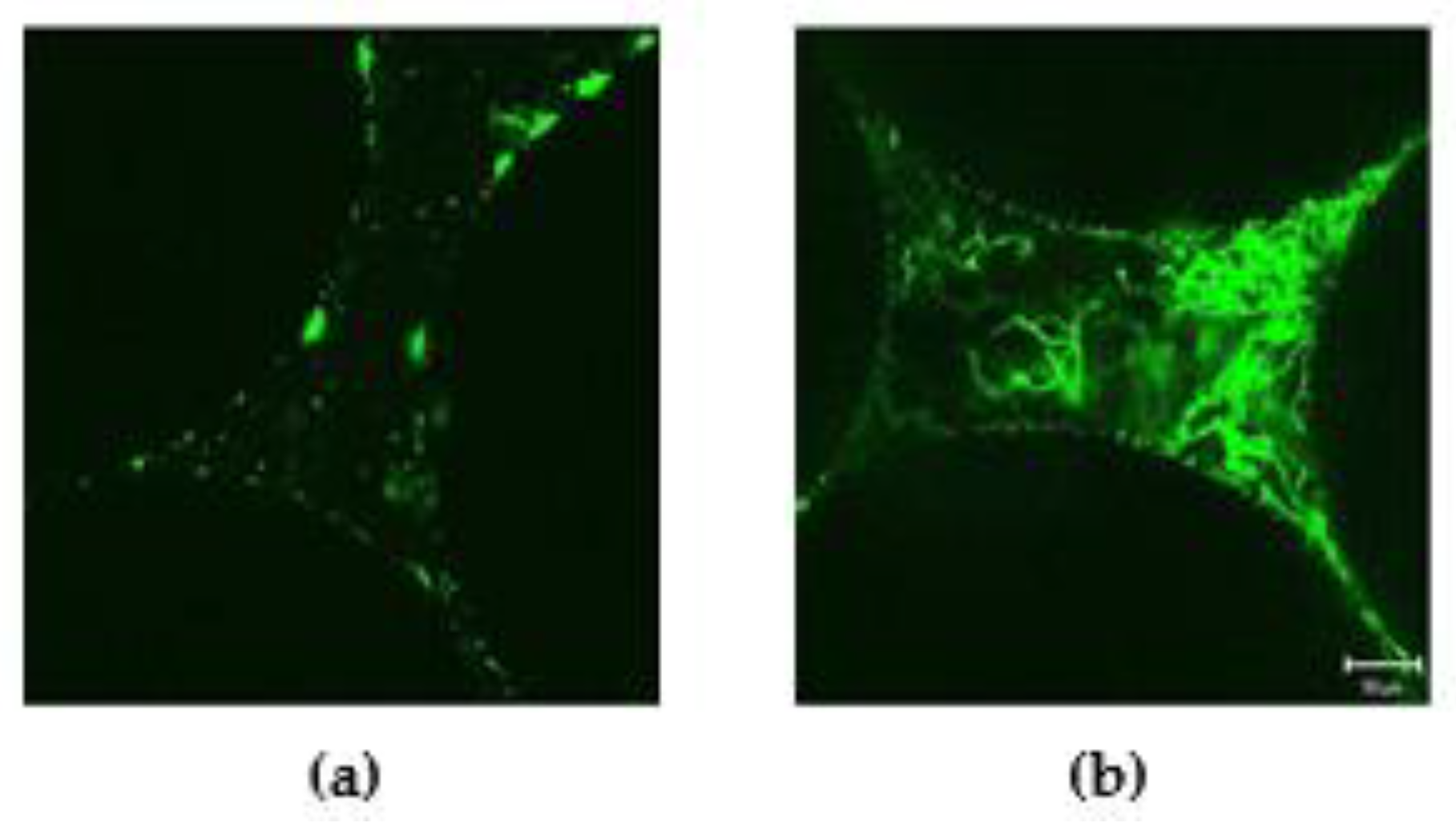
3.3. Confocal Laser Scanning Microscopy
3.4. Charge-Coupled Device (CCD) Camera
4. Mathematical Simulations of Biofilm Evolution in Porous Media
4.1. Effects of Biofilm Formation on Permeability
4.2. Interactions between Fluid Flow and Biofilm Development
4.3. Multispecies Biofilm Formation
5. Opportunities for In Situ Monitoring at Field Sites
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- O’Toole, G.; Kaplan, H.B.; Kolter, R. Biofilm formation as microbial development. Annu. Rev. Microbiol. 2000, 54, 49–79. [Google Scholar] [CrossRef] [PubMed]
- Stoodley, P.; Sauer, K.; Davies, D.G.; Costerton, J.W. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 2002, 56, 187–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flemming, H.C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef]
- Beer, D.D.; Stoodley, P.; Roe, F.; Lewandowski, Z. Effects of biofilm structures on oxygen distribution and mass transport. Biotechnol. Bioeng. 1993, 43, 1131–1138. [Google Scholar] [CrossRef] [PubMed]
- Palmer, R.J.J.; White, D.C. Developmental biology of biofilms: Implications for treatment and control. Trends Microbiol. 1997, 5, 435–440. [Google Scholar] [CrossRef]
- Shafahi, M.; Vafai, K. Synthesis of biofilm resistance characteristics against antibiotics. Int. J. Heat Mass Transf. 2010, 53, 2943–2950. [Google Scholar] [CrossRef]
- Ahmed, U.; Vafai, K. Analysis of biofilm growth in the presence of osmotic pressure and temperature effects. Int. J. Heat Mass Transf. 2012, 55, 5709–5721. [Google Scholar] [CrossRef] [Green Version]
- Hauser, M.; Vafai, K. Analysis of the multidimensional effects in biofilms. Int. J. Heat Mass Transf. 2013, 56, 340–349. [Google Scholar] [CrossRef] [Green Version]
- Vandevivere, P.; Baveye, P. Effect of bacterial extracellular polymers on the saturated hydraulic conductivity of sand columns. Appl. Environ. Microbiol. 1992, 58, 1690–1698. [Google Scholar] [CrossRef] [Green Version]
- Massol-Deyá, A.A.; Whallon, J.; Hickey, R.F.; Tiedje, J.M. Channel structures in aerobic biofilms of fixed-film reactors treating contaminated groundwater. Appl. Environ. Microbiol. 1995, 61, 769–777. [Google Scholar] [CrossRef] [Green Version]
- Pal, A.; Paul, A.K. Microbial extracellular polymeric substances central elements in heavy metal bioremediation. Indian J. Microbiol. 2008, 48, 49–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tufenkji, N. Modeling microbial transport in porous media: Traditional approaches and recent developments. Adv. Water Resour. 2007, 30, 1455–1469. [Google Scholar] [CrossRef] [Green Version]
- Rummel, C.D.; Jahnke, A.; Gorokhova, E.; Kühnel, D.; Schmitt-Jansen, M. Impacts of biofilm formation on the fate and potential effects of microplastic in the aquatic environment. Environ. Sci. Technol. Lett. 2017, 4, 258–267. [Google Scholar] [CrossRef] [Green Version]
- Guan, J.; Qi, K.; Wang, J.; Wang, W.; Wang, Z.; Lu, N.; Qu, J. Microplastics as an emerging anthropogenic vector of trace metals in freshwater: Significance of biofilms and comparison with natural substrates. Water Res. 2020, 184, 116205. [Google Scholar] [CrossRef]
- Shafahi, M.; Vafai, K. Biofilm affected characteristics of porous structures. Int. J. Heat Mass Transf. 2009, 52, 574–581. [Google Scholar] [CrossRef]
- Baveye, P.; Vandevivere, P.; Hoyle, B.L.; DeLeo, P.C.; de Lozada, D.S. Environmental impact and mechanisms of the biological clogging of saturated soils and squifer materials. Crit. Rev. Environ. Sci. Technol. 1998, 28, 123–191. [Google Scholar] [CrossRef] [Green Version]
- Taylor, S.W.; Jaffé, P.R. Biofilm growth and the related changes in the physical properties of a porous medium 1. Experimental investigation. Water Resour. Res. 1990, 26, 2153–2159. [Google Scholar]
- Bouwer, H. Artificial recharge of groundwater: Hydrogeology and engineering. Hydrogeol. J. 2002, 10, 121–142. [Google Scholar] [CrossRef] [Green Version]
- Davis, C.A.; Pyrak-Nolte, L.J.; Atekwana, E.A.; Werkema, D.D.; Haugen, M.E. Acoustic and electrical property changes due to microbial growth and biofilm formation in porous media. J. Geophys. Res. 2010, 115. [Google Scholar] [CrossRef] [Green Version]
- Rolland du Roscoat, S.; Martins, J.M.; Sechet, P.; Vince, E.; Latil, P.; Geindreau, C. Application of synchrotron X-ray microtomography for visualizing bacterial biofilms 3D microstructure in porous media. Biotechnol. Bioeng. 2014, 111, 1265–1271. [Google Scholar] [CrossRef]
- Trulear, M.G.; Characklis, W.G. Dynamics of biofilm processes. J. WPCF 1982, 54, 1288–1301. [Google Scholar]
- Stoodley, P.; Boyle, J.D.; DeBeer, D.; Lappin-Scott, H.M. Evolving perspectives of biofilm structure. Biofouling 1999, 14, 75–90. [Google Scholar] [CrossRef]
- Eighmy, T.T.; Maratea, D.; Bishop, P.L. Electron microscopic examination of wastewater biofilm formation and structural components. Appl. Environ. Microbiol. 1983, 45, 1921–1931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wijeyekoon, S.; Mino, T.; Satoh, H.; Matsuo, T. Effects of substrate loading rate on biofilm structure. Water Res. 2004, 38, 2479–2488. [Google Scholar] [CrossRef] [PubMed]
- Lecuyer, S.; Rusconi, R.; Shen, Y.; Forsyth, A.; Vlamakis, H.; Kolter, R.; Stone, H.A. Shear stress increases the residence time of adhesion of Pseudomonas aeruginosa. Biophys. J. 2011, 100, 341–350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stoodley, P.; Cargo, R.; Rupp, C.J.; Wilson, S.; Klapper, I. Biofilm material properties as related to shear-induced deformation and detachment phenomena. J. Ind. Microbiol. Biotechnol. 2002, 29, 361–367. [Google Scholar] [CrossRef]
- Wagner, M.; Taherzadeh, D.; Haisch, C.; Horn, H. Investigation of the mesoscale structure and volumetric features of biofilms using optical coherence tomography. Biotechnol. Bioeng. 2010, 107, 844–853. [Google Scholar] [CrossRef]
- Yang, J.; Cheng, S.; Li, C.; Sun, Y.; Huang, H. Shear stress affects biofilm structure and consequently current generation of bioanode in microbial electrochemical systems (MESs). Front. Microbiol. 2019, 10, 398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stoodley, P.; Dodds, I.; Boyle, J.D.; Lappin-Scott, H.M. Influence of hydrodynamics and nutrients on biofilm structure. J. Appl. Microbiol. 1999, 85, S19–S28. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, P.K.; Chand, D.V.; Chandra, J.; Anderson, J.M.; Ghannoum, M.A. Shear stress modulates the thickness and architecture of Candida albicans biofilms in a phase-dependent manner. Mycoses 2009, 52, 440–446. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.J.; Zhang, Z.; Bott, T.R. Effects of operating conditions on the adhesive strength of Pseudomonas fluorescens biofilms in tubes. Colloids Surf. B Biointerfaces 2005, 43, 61–71. [Google Scholar] [CrossRef]
- Wagner, M.; Manz, B.; Volke, F.; Neu, T.R.; Horn, H. Online assessment of biofilm development, sloughing and forced detachment in tube reactor by means of magnetic resonance microscopy. Biotechnol. Bioeng. 2010, 107, 172–181. [Google Scholar] [CrossRef] [PubMed]
- Gjermansen, M.; Ragas, P.; Sternberg, C.; Molin, S.; Tolker-Nielsen, T. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms. Environ. Microbiol. 2005, 7, 894–906. [Google Scholar] [CrossRef]
- Guillemot, G.; Vaca-Medina, G.; Martin-Yken, H.; Vernhet, A.; Schmitz, P.; Mercier-Bonin, M. Shear-flow induced detachment of Saccharomyces cerevisiae from stainless steel: Influence of yeast and solid surface properties. Colloids Surf. B Biointerfaces 2006, 49, 126–135. [Google Scholar] [CrossRef]
- Gjermansen, M.; Nilsson, M.; Yang, L.; Tolker-Nielsen, T. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms: Genetic elements and molecular mechanisms. Mol. Microbiol. 2010, 75, 815–826. [Google Scholar] [CrossRef]
- Qin, C.-Z.; Hassanizadeh, S.M. Pore-network modeling of solute transport and biofilm growth in porous media. Transp. Porous Media 2015, 110, 345–367. [Google Scholar] [CrossRef] [Green Version]
- Cunningham, A.B.; Characklls, W.G.; Abedeen, F.; Crawford, D. Influence of biofilm accumulation on porous media hydrodynamics. Environ. Sci. Technol. 1991, 25, 1305–1311. [Google Scholar] [CrossRef]
- Kirk, M.F.; Santillan, E.F.U.; McGrath, L.K.; Altman, S.J. Variation in hydraulic conductivity with decreasing pH in a biologically-clogged porous medium. Int. J. Greenh. Gas. Control 2012, 11, 133–140. [Google Scholar] [CrossRef]
- Seymour, J.D.; Gage, J.P.; Codd, S.L.; Gerlach, R. Anomalous fluid transport in porous media induced by biofilm growth. Phys. Rev. Lett. 2004, 93, 198103. [Google Scholar] [CrossRef]
- Stoodley, P.; Debeer, D.; Lewandowski, Z. Liquid flow in biofilm systems. Appl. Environ. Microbiol. 1994, 60, 2711–2716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.; Lewandowski, Z. Measurement of local mass transfer coefficient in biofilms. Biotechnol. Bioeng. 1995, 48, 737–744. [Google Scholar] [CrossRef] [PubMed]
- Tolker-Nielsen, T.; Brinch, U.C.; Ragas, P.C.; Andersen, J.B.; Jacobsen, C.S.; Molin, S. Development and dynamics of Pseudomonas sp. biofilms. J. Bacteriol. 2000, 182, 6482–6489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manz, B.; Volke, F.; Goll, D.; Horn, H. Measuring local flow velocities and biofilm structure in biofilm systems with magnetic resonance imaging (MRI). Biotechnol. Bioeng. 2003, 84, 424–432. [Google Scholar] [CrossRef] [PubMed]
- Klausen, M.; Aaes-Jorgensen, A.; Molin, S.; Tolker-Nielsen, T. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol. Microbiol. 2003, 50, 61–68. [Google Scholar] [CrossRef] [Green Version]
- Seymour, J.D.; Codd, S.L.; Gjersing, E.L.; Stewart, P.S. Magnetic resonance microscopy of biofilm structure and impact on transport in a capillary bioreactor. J. Magn. Reson. 2004, 167, 322–327. [Google Scholar] [CrossRef] [PubMed]
- Gjersing, E.L.; Codd, S.L.; Seymour, J.D.; Stewart, P.S. Magnetic resonance microscopy analysis of advective transport in a biofilm reactor. Biotechnol. Bioeng. 2005, 89, 822–834. [Google Scholar] [CrossRef] [PubMed]
- Vandevivere, P.; Baveye, P. Saturated hydraulic conductivity reduction caused by aerobic bacteria in sand columns. Soil Sci. Soc. Am. J. 1992, 56, 1–13. [Google Scholar] [CrossRef]
- Vandevivere, P.; Baveye, P. Relationship between transport of bacteria and their clogging efficiency in sand columns. Appl. Environ. Microbiol. 1992, 58, 2523–2530. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.-S.; Fogler, H.S. Biomass evolution in porous media and its effects on permeability under starvation conditions. Biotechnol. Bioeng. 2000, 69, 47–56. [Google Scholar] [CrossRef]
- Davis, C.A.; Pyrak-Nolte, L.J.; Atekwana, E.A.; Werkema, D.D.; Haugen, M.E. Microbial-induced heterogeneity in the acoustic properties of porous media. Geophys. Res. Lett. 2009, 36. [Google Scholar] [CrossRef] [Green Version]
- Stewart, T.L.; Fogler, H.S. Biomass plug development and propagation in porous media. Biotechnol. Bioeng. 2000, 72, 353–363. [Google Scholar] [CrossRef] [Green Version]
- Stewart, T.L.; Scott Fogler, H. Pore-scale investigation of biomass plug development and propagation in porous media. Biotechnol. Bioeng. 2002, 77, 577–588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunsmore, B.C.; Bass, C.J.; Lappin-Scott, H.M. A novel approach to investigate biofilm accumulation and bacterial transport in porous matrices. Environ. Microbiol. 2004, 6, 183–187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iltis, G.C.; Armstrong, R.T.; Jansik, D.P.; Wood, B.D.; Wildenschild, D. Imaging biofilm architecture within porous media using synchrotron-based X-ray computed microtomography. Water Resour. Res. 2011, 47. [Google Scholar] [CrossRef]
- Davit, Y.; Iltis, G.; Debenest, G.; Veran-Tissoires, S.; Wildenschild, D.; Gerino, M.; Quintard, M. Imaging biofilm in porous media using X-ray computed microtomography. J. Microsc. 2011, 242, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Carrel, M.; Beltran, M.A.; Morales, V.L.; Derlon, N.; Morgenroth, E.; Kaufmann, R.; Holzner, M. Biofilm imaging in porous media by laboratory X-ray tomography: Combining a non-destructive contrast agent with propagation-based phase-contrast imaging tools. PLoS ONE 2017, 12, e0180374. [Google Scholar] [CrossRef] [Green Version]
- Carrel, M.; Morales, V.L.; Beltran, M.A.; Derlon, N.; Kaufmann, R.; Morgenroth, E.; Holzner, M. Biofilms in 3D porous media: Delineating the influence of the pore network geometry, flow and mass transfer on biofilm development. Water Res. 2018, 134, 280–291. [Google Scholar] [CrossRef] [Green Version]
- Hoskins, B.C.; Fevang, L.; Majors, P.D.; Sharma, M.M.; Georgiou, G. Selective imaging of biofilms in porous media by NMR relaxation. J. Magn. Reson. 1999, 139, 67–73. [Google Scholar] [CrossRef]
- Seymour, J.D.; Gage, J.P.; Codd, S.L.; Gerlach, R. Magnetic resonance microscopy of biofouling induced scale dependent transport in porous media. Adv. Water Resour. 2007, 30, 1408–1420. [Google Scholar] [CrossRef]
- Codd, S.L.; Vogt, S.J.; Hornemann, J.A.; Phillips, A.J.; Maneval, J.E.; Romanenko, K.R.; Hansen, L.; Cunningham, A.B.; Seymour, J.D. NMR relaxation measurements of biofouling in model and geological porous media. Org. Geochem. 2011, 42, 965–971. [Google Scholar] [CrossRef] [Green Version]
- Kirkland, C.M.; Herrling, M.P.; Hiebert, R.; Bender, A.T.; Grunewald, E.; Walsh, D.O.; Codd, S.L. In situ detection of subsurface biofilm using low-field NMR: A field study. Environ. Sci. Technol. 2015, 49, 11045–11052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.W.; Choi, H.; Pachepsky, Y.A. Biofilm morphology as related to the porous media clogging. Water Res. 2010, 44, 1193–1201. [Google Scholar] [CrossRef]
- Kone, T.; Golfier, F.; Orgogozo, L.; Oltéan, C.; Lefèvre, E.; Block, J.C.; Buès, M.A. Impact of biofilm-induced heterogeneities on solute transport in porous media. Water Resour. Res. 2014, 50, 9103–9119. [Google Scholar] [CrossRef]
- Nakashima, Y.; Nakano, T.; Nakamura, K.; Uesugi, K.; Tsuchiyama, A.; Ikeda, S. Three-dimensional diffusion of non-sorbing species in porous sandstone: Computer simulation based on X-ray microtomography using synchrotron radiation. J. Contam. Hydrol. 2004, 74, 253–264. [Google Scholar] [CrossRef] [Green Version]
- Werth, C.J.; Zhang, C.; Brusseau, M.L.; Oostrom, M.; Baumann, T. A review of non-invasive imaging methods and applications in contaminant hydrogeology research. J. Contam. Hydrol. 2010, 113, 1–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- San José Martínez, F.; Muñoz Ortega, F.J.; Caniego Monreal, F.J.; Peregrina, F. Morphological functions with parallel sets for the pore space of X-ray CT images of soil columns. Pure Appl. Geophys. 2014, 173, 995–1009. [Google Scholar] [CrossRef]
- Zhou, H.; Mooney, S.J.; Peng, X. Bimodal soil pore structure investigated by a combined soil water retention curve and X-Ray computed tomography approach. Soil Sci. Soc. Am. J. 2017, 81, 1270–1278. [Google Scholar] [CrossRef]
- Kutsovsky, Y.E.; Scriven, L.E.; Davis, H.T.; Hammer, B.E. NMR imaging of velocity profiles and velocity distributions in bead packs. Phys. Fluids 1996, 8, 863–871. [Google Scholar] [CrossRef]
- Dijk, P.; Berkowitz, B.; Bendel, P. Investigation of flow in water-saturated rock fractures using nuclear magnetic resonance imaging (NMRI). Water Resour. Res. 1999, 35, 347–360. [Google Scholar] [CrossRef]
- Neu, T.R.; Manz, B.; Volke, F.; Dynes, J.J.; Hitchcock, A.P.; Lawrence, J.R. Advanced imaging techniques for assessment of structure, composition and function in biofilm systems. FEMS Microbiol. Ecol. 2010, 72, 1–21. [Google Scholar] [CrossRef]
- Eberl, H.J.; Picioreanu, C.; Heijnen, J.J.; Loosdrecht, M.C.M.V. A three-dimensional numerical study on the correlation of spatial structure, hydrodynamic conditions, and mass transfer and conversion in biofilms. Chem. Eng. Sci. 2000, 55, 6209–6222. [Google Scholar] [CrossRef]
- Stoodley, P.; Yang, S.; Lappin-Scott, H.; Lewandowski, Z. Relationship between mass transfer coefficient and liquid flow velocity in heterogenous biofilms using microelectrodes and confocal microscopy. Biotechnol. Bioeng. 1997, 56, 681–688. [Google Scholar] [CrossRef]
- Stewart, P.S.; Peyton, B.M.; Drury, W.J.; Murga, R. Quantitative observations of heterogeneities in Pseudomonas aeruginosa biofilms. Appl.Environ. Microbiol. 1993, 59, 327–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murga, R.; Stewart, P.; Daly, D. Quantitative analysis of biofilm thickness variability. Biotechnol. Bioeng. 1994, 45, 503–510. [Google Scholar] [CrossRef]
- Heydorn, A.; Nielsen, A.T.; Hentzer, M.; Sternberg, C.; Givskov, M.; Ersbøll, B.K.; Molin, S. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 2000, 146, 2395–2407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hentzer, M.; Teitzel, G.M.; Balzer, G.J.; Heydorn, A.; Molin, S.; Givskov, M.; Parsek, M.R. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J. Bacteriol. 2001, 183, 5395–5401. [Google Scholar] [CrossRef] [Green Version]
- Garny, K.; Horn, H.; Neu, T.R. Interaction between biofilm development, structure and detachment in rotating annular reactors. Bioprocess Biosyst. Eng. 2008, 31, 619–629. [Google Scholar] [CrossRef]
- Kim, S.R.; Oh, H.S.; Jo, S.J.; Yeon, K.M.; Lee, C.H.; Lim, D.J.; Lee, C.H.; Lee, J.K. Biofouling control with bead-entrapped quorum quenching bacteria in membrane bioreactors: Physical and biological effects. Environ. Sci. Technol. 2013, 47, 836–842. [Google Scholar] [CrossRef]
- Rodriguez-Melcon, C.; Alonso-Hernando, A.; Riesco-Pelaez, F.; Garcia-Fernandez, C.; Alonso-Calleja, C.; Capita, R. Biovolume and spatial distribution of foodborne Gram-negative and Gram-positive pathogenic bacteria in mono- and dual-species biofilms. Food Microbiol. 2021, 94, 103616. [Google Scholar] [CrossRef]
- Knutson, C.E.; Werth, C.J.; Valocchi, A.J. Pore-scale simulation of biomass growth along the transverse mixing zone of a model two-dimensional porous medium. Water Resour. Res. 2005, 41. [Google Scholar] [CrossRef]
- Zhang, C.; Kang, Q.; Wang, X.; Zilles, J.L.; Muller, R.H.; Werth, C.J. Effects of pore-scale heterogeneity and transverse mixing on bacterial growth in porous media. Environ. Sci. Technol. 2010, 44, 3085–3092. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Valocchi, A.J.; Werth, C.J.; Liu, H. An improved pore-scale biofilm model and comparison with a microfluidic flow cell experiment. Water Resour. Res. 2013, 49, 8370–8382. [Google Scholar] [CrossRef]
- Tang, Y.; Liu, H. Modeling multidimensional and multispecies biofilms in porous media. Biotechnol. Bioeng. 2017, 114, 1679–1687. [Google Scholar] [CrossRef] [PubMed]
- Aghajani Delavar, M.; Wang, J. Pore-scale modeling of competition and cooperation of multispecies biofilms for nutrients in changing environments. AIChE J. 2020, 66. [Google Scholar] [CrossRef]
- Stewart, T.L.; Kim, D.-S. Modeling of biomass-plug development and propagation in porous media. Biochem. Eng. J. 2004, 17, 107–119. [Google Scholar] [CrossRef]
- Kapellos, G.E.; Alexiou, T.S.; Payatakes, A.C. Hierarchical simulator of biofilm growth and dynamics in granular porous materials. Adv. Water Resour. 2007, 30, 1648–1667. [Google Scholar] [CrossRef]
- Golfier, F.; Wood, B.D.; Orgogozo, L.; Quintard, M.; Buès, M. Biofilms in porous media: Development of macroscopic transport equations via volume averaging with closure for local mass equilibrium conditions. Adv. Water Resour. 2009, 32, 463–485. [Google Scholar] [CrossRef]
- Pintelon, T.R.; Graf von der Schulenburg, D.A.; Johns, M.L. Towards optimum permeability reduction in porous media using biofilm growth simulations. Biotechnol. Bioeng. 2009, 103, 767–779. [Google Scholar] [CrossRef]
- Graf von der Schulenburg, D.A.; Pintelon, T.R.R.; Picioreanu, C.; Van Loosdrecht, M.C.M.; Johns, M.L. Three-dimensional simulations of biofilm growth in porous media. AIChE J. 2009, 55, 494–504. [Google Scholar] [CrossRef]
- Ebigbo, A.; Helmig, R.; Cunningham, A.B.; Class, H.; Gerlach, R. Modelling biofilm growth in the presence of carbon dioxide and water flow in the subsurface. Adv. Water Resour. 2010, 33, 762–781. [Google Scholar] [CrossRef]
- Deng, W.; Cardenas, M.B.; Kirk, M.F.; Altman, S.J.; Bennett, P.C. Effect of permeable biofilm on micro- and macro-scale flow and transport in bioclogged pores. Environ. Sci. Technol. 2013, 47, 11092–11098. [Google Scholar] [CrossRef] [PubMed]
- Picioreanu, C.; Loosdrecht, M.C.M.V. Use of mathematical modelling to study biofilm development and morphology. In Biofilms in Medicine, Industry and Environmental Biotechnology: Characteristics, Analysis and Control; Lens, P., Moran, A.P., Mahony, T., Stoodley, P., O’Flaherty, V., Eds.; IWA: London, UK, 2003; pp. 413–438. [Google Scholar]
- Loosdrecht, M.C.M.V.; Heijnen, J.J.; Eberl, H.; Kreft, J.; Picioreanu, C. Mathematical modelling of biofilm structures. Anton. Van Leeuw. Int. J. G 2002, 81, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Mattei, M.R.; Frunzo, L.; D’Acunto, B.; Pechaud, Y.; Pirozzi, F.; Esposito, G. Continuum and discrete approach in modeling biofilm development and structure: A review. J. Math. Biol. 2018, 76, 945–1003. [Google Scholar] [CrossRef] [Green Version]
- Hermanowicz, S.W. A simple 2D biofilm model yields a variety of morphological features. Math. Biosci. 2000, 169, 1–14. [Google Scholar] [CrossRef]
- Hunt, S.M.; Werner, E.M.; Huang, B.; Hamilton, M.A.; Stewart, P.S. Hypothesis for the role of nutrient starvation in biofilm detachment. Appl. Environ. Microbiol. 2004, 70, 7418–7425. [Google Scholar] [CrossRef] [Green Version]
- Acemel, R.D.; Govantes, F.; Cuetos, A. Computer simulation study of early bacterial biofilm development. Sci. Rep. 2018, 8, 5340. [Google Scholar] [CrossRef]
- Luna, E.; Dominguez-Zacarias, G.; Ferreira, C.P.; Velasco-Hernandez, J.X. Detachment and diffusive-convective transport in an evolving heterogeneous two-dimensional biofilm hybrid model. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2004, 70, 061909. [Google Scholar] [CrossRef] [Green Version]
- Klapper, I.; Rupp, C.J.; Cargo, R.; Purvedorj, B.; Stoodley, P. Viscoelastic fluid description of bacterial biofilm material properties. Biotechnol. Bioeng. 2002, 80, 289–296. [Google Scholar] [CrossRef]
- Chambless, J.D.; Stewart, P.S. A three-dimensional computer model analysis of three hypothetical biofilm detachment mechanisms. Biotechnol. Bioeng. 2007, 97, 1573–1584. [Google Scholar] [CrossRef]
- Cunningham, J.A.; Mendoza-Sanchez, I. Equivalence of two models for biodegradation during contaminant transport in groundwater. Water Resour. Res. 2006, 42. [Google Scholar] [CrossRef] [Green Version]
- D’Acunto, B.; Esposito, G.; Frunzo, L.; Mattei, M.R.; Pirozzi, F. Mathematical modeling of heavy metal biosorption in multispecies biofilms. J. Environ. Eng. 2016, 142. [Google Scholar] [CrossRef]
- Picioreanu, C.; Loosdrecht, M.C.M.V.; Heijnen, J.J. Effect of diffusive and convective substrate transport on biofilm structure formation: A two-dimensional modeling study. Biotechnol. Bioeng. 2000, 69, 504–515. [Google Scholar] [CrossRef]
- Taherzadeh, D.; Picioreanu, C.; Horn, H. Mass transfer enhancement in moving biofilm structures. Biophys. J. 2012, 102, 1483–1492. [Google Scholar] [CrossRef] [Green Version]
- Pintelon, T.R.; Picioreanu, C.; Loosdrecht, M.C.; Johns, M.L. The effect of biofilm permeability on bio-clogging of porous media. Biotechnol. Bioeng. 2012, 109, 1031–1042. [Google Scholar] [CrossRef]
- Bottero, S.; Storck, T.; Heimovaara, T.J.; van Loosdrecht, M.C.; Enzien, M.V.; Picioreanu, C. Biofilm development and the dynamics of preferential flow paths in porous media. Biofouling 2013, 29, 1069–1086. [Google Scholar] [CrossRef]
- Komlos, J.; Cunningham, A.B.; Camper, A.K.; Sharp, R.R. Biofilm barriers to contain and degrade dissolved trichloroethylene. Environ. Prog. 2004, 23, 69–77. [Google Scholar] [CrossRef]
- Mitchell, A.C.; Phillips, A.J.; Hiebert, R.; Gerlach, R.; Spangler, L.H.; Cunningham, A.B. Biofilm enhanced geologic sequestration of supercritical CO2. Int. J. Greenh. Gas. Con. 2009, 3, 90–99. [Google Scholar] [CrossRef] [Green Version]
- Abdel Aal, G.Z.; Atekwana, E.A. Spectral induced polarization (SIP) response of biodegraded oil in porous media. Geophys. J. Int. 2014, 196, 804–817. [Google Scholar] [CrossRef] [Green Version]
- Lu, L.; Huggins, T.; Jin, S.; Zuo, Y.; Ren, Z.J. Microbial metabolism and community structure in response to bioelectrochemically enhanced remediation of petroleum hydrocarbon-contaminated soil. Environ. Sci. Technol. 2014, 48, 4021–4029. [Google Scholar] [CrossRef]
- Davis, C.A.; Atekwana, E.; Atekwana, E.; Slater, L.D.; Rossbach, S.; Mormile, M.R. Microbial growth and biofilm formation in geologic media is detected with complex conductivity measurements. Geophys. Res. Lett. 2006, 33, L18403. [Google Scholar] [CrossRef]
- Abdel Aal, G.Z.; Atekwana, E.A.; Atekwana, E.A. Effect of bioclogging in porous media on complex conductivity signatures. J. Geophys. Res. 2010, 115. [Google Scholar] [CrossRef] [Green Version]
- Godefroy, S.; Korb, J.P.; Fleury, M.; Bryant, R.G. Surface nuclear magnetic relaxation and dynamics of water and oil in macroporous media. Phys. Rev. E Stat. Nonlinear Soft Matter Phys. 2001, 64, 021605. [Google Scholar] [CrossRef]
- Heij, E.J.L.; Kerkhof, P.J.A.M.; Kopinga, K.; Pel, L. Determining expression porosity profiles during filtration and of sewage sludge by NMR imaging. AIChE J. 1996, 42, 953–959. [Google Scholar] [CrossRef] [Green Version]
- Bryar, T.R.; Knight, R.J. NMR relaxation measurements to quantify immiscible organic contaminants in sediments. Water Resour. Res. 2008, 44. [Google Scholar] [CrossRef]
- Dlubac, K.; Knight, R.; Song, Y.-Q.; Bachman, N.; Grau, B.; Cannia, J.; Williams, J. Use of NMR logging to obtain estimates of hydraulic conductivity in the High Plains aquifer, Nebraska, USA. Water Resour. Res. 2013, 49, 1871–1886. [Google Scholar] [CrossRef] [Green Version]
- Parsekian, A.D.; Grosse, G.; Walbrecker, J.O.; Müller-Petke, M.; Keating, K.; Liu, L.; Jones, B.M.; Knight, R. Detecting unfrozen sediments below thermokarst lakes with surface nuclear magnetic resonance. Geophys. Res. Lett. 2013, 40, 535–540. [Google Scholar] [CrossRef]
- Sanderlin, A.B.; Vogt, S.J.; Grunewald, E.; Bergin, B.A.; Codd, S.L. Biofilm detection in natural unconsolidated porous media using a low-field magnetic resonance system. Environ. Sci. Technol. 2013, 47, 987–992. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Method | Porous Media | Grain Size (μm) |
|---|---|---|
| X-ray | Glass beads [54] | >1000 |
| Expanded polystyrene beads [55] | 500–1500 | |
| Clay beads [20] | >1000 | |
| Nafion pellets [56] | 2500 | |
| Transparent Nafion pellets [57] | 2500 | |
| NMR | Glass beads [58] | 1000–1500 |
| Monodisperse beads [39] | 241 | |
| Monodisperse beads [59] | 241 | |
| Glass beads [60] | 100 | |
| Soil [61] | - | |
| CLSM | Glass beads [62] | 425–600 |
| CCD | Silica sand [63] | 210–297 |
| Contrast Agent | Disadvantages |
|---|---|
| Hollow silver-coated microspheres [54] | Toxic Limited surface attachment |
| Mixture of barium sulfate and potassium iodide [55] | Toxic Fast sedimentation Heterogeneous distribution |
| 1-Chloronaphtalene [20] | Toxic Immiscible with water |
| Iron sulfate [56] | Less soluble Form colloids |
| Barium sulfate [57] | Toxic Fast sedimentation Heterogeneous distribution |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Xu, A.; Lv, X.; Wang, Q.; Feng, C.; Lin, J. Non-Invasive Measurement, Mathematical Simulation and In Situ Detection of Biofilm Evolution in Porous Media: A Review. Appl. Sci. 2021, 11, 1391. https://doi.org/10.3390/app11041391
Zhang Y, Xu A, Lv X, Wang Q, Feng C, Lin J. Non-Invasive Measurement, Mathematical Simulation and In Situ Detection of Biofilm Evolution in Porous Media: A Review. Applied Sciences. 2021; 11(4):1391. https://doi.org/10.3390/app11041391
Chicago/Turabian StyleZhang, Yajun, Aoshu Xu, Xin Lv, Qian Wang, Caihui Feng, and Jun Lin. 2021. "Non-Invasive Measurement, Mathematical Simulation and In Situ Detection of Biofilm Evolution in Porous Media: A Review" Applied Sciences 11, no. 4: 1391. https://doi.org/10.3390/app11041391
APA StyleZhang, Y., Xu, A., Lv, X., Wang, Q., Feng, C., & Lin, J. (2021). Non-Invasive Measurement, Mathematical Simulation and In Situ Detection of Biofilm Evolution in Porous Media: A Review. Applied Sciences, 11(4), 1391. https://doi.org/10.3390/app11041391






