A Serum Metabolomic Signature for the Detection and Grading of Bladder Cancer
Abstract
:Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Population and Study Design
2.2. Data and Sample Collection
2.3. Histopathological Analysis
2.4. Metabolome Analysis by GC-MS
2.5. Statistical Analysis
2.5.1. Classification Models
2.5.2. Ensemble Machine Learning Score (EML-Score)
2.6. Relevant Metabolites
2.6.1. Variables Important in Projection (VIP)
2.6.2. Volcano Plot
2.6.3. Genetic Algorithm
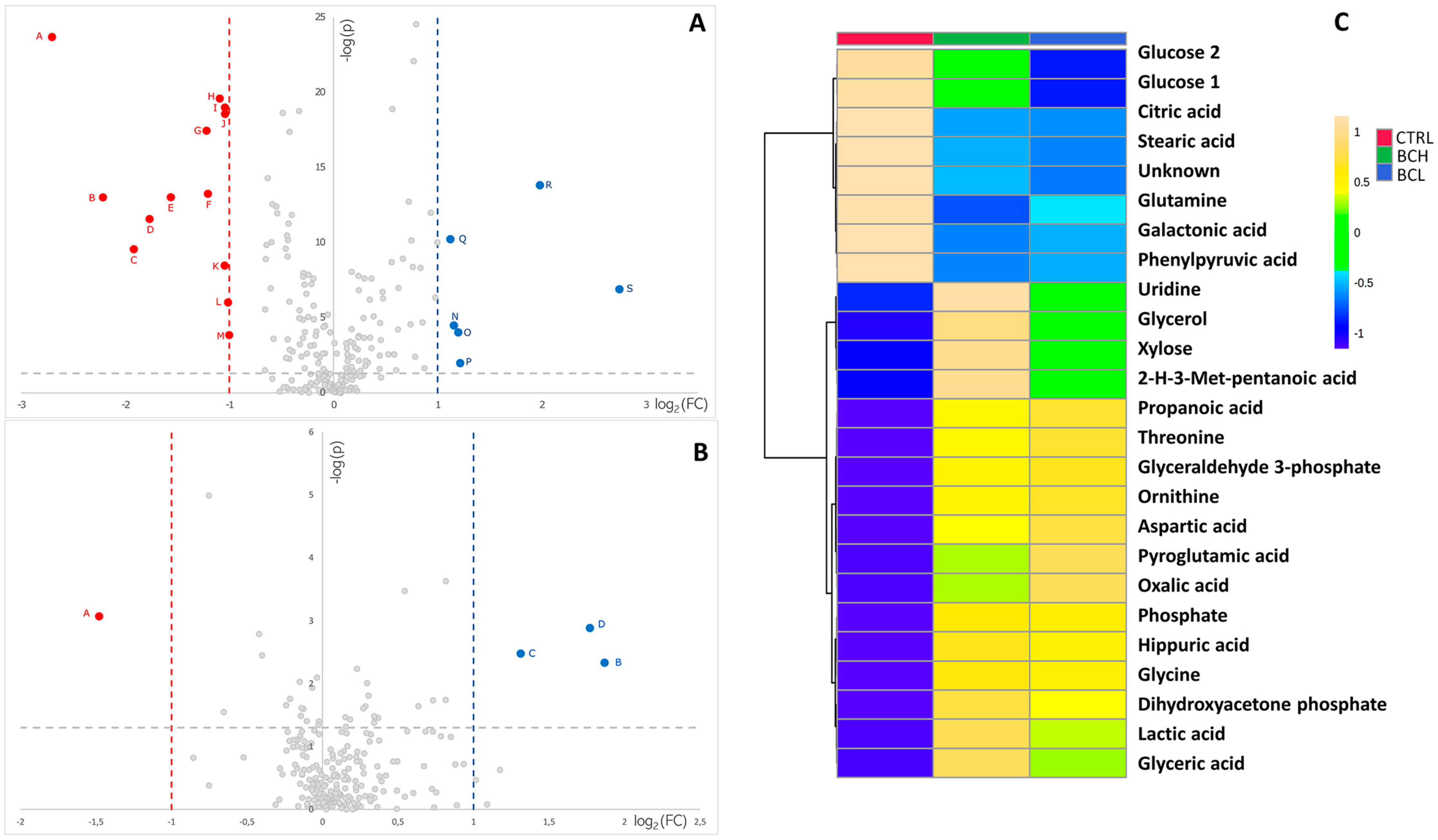
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Dyba, T.; Randi, G.; Bettio, M.; Gavin, A.; Visser, O.; Bray, F. Cancer Incidence and Mortality Patterns in Europe: Estimates for 40 Countries and 25 Major Cancers in 2018. Eur. J. Cancer 2018, 103, 356–387. [Google Scholar] [CrossRef]
- Bladder Cancer: Diagnosis and Management of Bladder Cancer. BJU Int. 2017, 120, 755–765. [CrossRef] [Green Version]
- Burger, M.; Catto, J.W.F.; Dalbagni, G.; Grossman, H.B.; Herr, H.; Karakiewicz, P.; Kassouf, W.; Kiemeney, L.A.; La Vecchia, C.; Shariat, S.; et al. Epidemiology and Risk Factors of Urothelial Bladder Cancer. Eur. Urol. 2013, 63, 234–241. [Google Scholar] [CrossRef]
- Daneshmand, S. Epidemiology and Risk Factors of Urothelial (Transitional Cell) Carcinoma of the Bladder. UpToDate. 2016. Available online: http://www.uptodate.com/contents/epidemiology-and-risk-factors-of-urothelial-transitional-cell-carcinoma-of-the-bladder (accessed on 6 July 2017).
- Abraham, J.; Gulley, J.L.; Allegra, C.J. The Bethesda Handbook of Clinical Oncology; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2012. [Google Scholar]
- Redondo-Gonzalez, E.; de Castro, L.N.; Moreno-Sierra, J.; Maestro de las Casas, M.L.; Vera-Gonzalez, V.; Ferrari, D.G.; Corchado, J.M. Bladder Carcinoma Data with Clinical Risk Factors and Molecular Markers: A Cluster Analysis. BioMed Res. Int. 2015, 2015, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El Mohsen, M.; Shelbaia, A.; El, S.G. Sequential Chemoimmunotherapy Using Mitomycin Followed by Bacillus Calmette-Guerin (MCC+ BCG) versus Single-Agent Immunotherapy (BCG) for Recurrent Superficial Bladder Tumors. UroToday Int. J. 2010, 3. [Google Scholar] [CrossRef]
- Park, J.C.; Citrin, D.E.; Agarwal, P.K.; Apolo, A.B. Multimodal Management of Muscle Invasive Bladder Cancer. Curr. Probl. Cancer 2014, 38, 80–108. [Google Scholar] [CrossRef] [Green Version]
- Yokomizo, A.; Kanimoto, Y.; Okamura, T.; Ozono, S.; Koga, H.; Iwamura, M.; Tanaka, H.; Takahashi, S.; Tsushima, T.; Kanayama, H.; et al. Randomized Controlled Study of the Efficacy, Safety and Quality of Life with Low Dose Bacillus Calmette-Guérin Instillation Therapy for Nonmuscle Invasive Bladder Cancer. J. Urol. 2016, 195, 41–46. [Google Scholar] [CrossRef]
- Stein, J.P.; Lieskovsky, G.; Cote, R.; Groshen, S.; Feng, A.-C.; Boyd, S.; Skinner, E.; Bochner, B.; Thangathurai, D.; Mikhail, M.; et al. Radical Cystectomy in the Treatment of Invasive Bladder Cancer: Long-Term Results in 1054 Patients. J. Clin. Oncol. 2001, 19, 666–675. [Google Scholar] [CrossRef] [PubMed]
- Grossman, H.B.; Natale, R.B.; Tangen, C.M.; Speights, V.O.; Vogelzang, N.J.; Trump, D.L.; deVere White, R.W.; Sarosdy, M.F.; Wood, D.P., Jr.; Raghavan, D. Neoadjuvant Chemotherapy plus Cystectomy Compared with Cystectomy Alone for Locally Advanced Bladder Cancer. N. Engl. J. Med. 2003, 349, 859–866. [Google Scholar] [CrossRef] [PubMed]
- Kamoun, A.; de Reyniès, A.; Allory, Y.; Sjödahl, G.; Robertson, A.G.; Seiler, R.; Hoadley, K.A.; Groeneveld, C.S.; Al-Ahmadie, H.; Choi, W.; et al. A Consensus Molecular Classification of Muscle-Invasive Bladder Cancer. Eur. Urol. 2020, 77, 420–433. [Google Scholar] [CrossRef] [PubMed]
- Caputo, A.; D’Antonio, A.; Memoli, D.; Sabbatino, F.; Altieri, V.; Zeppa, P. Ki67 in Gleason Pattern 3 as a Marker of the Presence of Higher-Grade Prostate Cancer. Appl. Immunohistochem. Mol. Morphol. 2021, 29, 112–117. [Google Scholar] [CrossRef]
- Troisi, J.; Sarno, L.; Landolfi, A.; Scala, G.; Martinelli, P.; Venturella, R.; Di Cello, A.; Zullo, F.; Guida, M. Metabolomic Signature of Endometrial Cancer. J. Proteome Res. 2018, 17, 804–812. [Google Scholar] [CrossRef] [PubMed]
- Troisi, J.; Sarno, L.; Martinelli, P.; Di Carlo, C.; Landolfi, A.; Scala, G.; Rinaldi, M.; D’Alessandro, P.; Ciccone, C.; Guida, M. A Metabolomics-Based Approach for Non-Invasive Diagnosis of Chromosomal Anomalies. Metabolomics 2017, 13, 140. [Google Scholar] [CrossRef]
- Troisi, J.; Landolfi, A.; Sarno, L.; Richards, S.; Symes, S.; Adair, D.; Ciccone, C.; Scala, G.; Martinelli, P.; Guida, M. A Metabolomics-Based Approach for Non-Invasive Screening of Fetal Central Nervous System Anomalies. Metabolomics 2018, 14, 77. [Google Scholar] [CrossRef]
- Troisi, J.; Raffone, A.; Travaglino, A.; Belli, G.; Belli, C.; Anand, S.; Giugliano, L.; Cavallo, P.; Scala, G.; Symes, S.; et al. Development and Validation of a Serum Metabolomic Signature for Endometrial Cancer Screening in Postmenopausal Women. JAMA Netw. Open 2020, 3, e2018327. [Google Scholar] [CrossRef] [PubMed]
- Troisi, J.; Cavallo, P.; Richards, S.; Symes, S.; Colucci, A.; Sarno, L.; Landolfi, A.; Scala, G.; Adair, D.; Ciccone, C.; et al. Non-Invasive Screening for Congenital Heart Defects Using a Serum Metabolomics Approach. Prenat. Diagn. 2021. [Google Scholar] [CrossRef] [PubMed]
- R Development CORE TEAM, R. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2008. [Google Scholar]
- Domingos, P. MetaCost: A General Method for Making Classifiers Cost-Sensitive. In Proceedings of the 5th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Diego, CA, USA, 15–18 August 1999; pp. 155–164. [Google Scholar]
- Rubingh, C.M.; Bijlsma, S.; Derks, E.P.P.A.; Bobeldijk, I.; Verheij, E.R.; Kochhar, S.; Smilde, A.K. Assessing the Performance of Statistical Validation Tools for Megavariate Metabolomics Data. Metabolomics 2006, 2, 53–61. [Google Scholar] [CrossRef] [Green Version]
- Westerhuis, J.A.; Hoefsloot, H.C.J.; Smit, S.; Vis, D.J.; Smilde, A.K.; van Velzen, E.J.J.; van Duijnhoven, J.P.M.; van Dorsten, F.A. Assessment of PLSDA Cross Validation. Metabolomics 2008, 4, 81–89. [Google Scholar] [CrossRef] [Green Version]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vazquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The Human Metabolome Database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef] [PubMed]
- Karnovsky, A.; Weymouth, T.; Hull, T.; Tarcea, V.G.; Scardoni, G.; Laudanna, C.; Sartor, M.A.; Stringer, K.A.; Jagadish, H.V.; Burant, C.; et al. Metscape 2 Bioinformatics Tool for the Analysis and Visualization of Metabolomics and Gene Expression Data. Bioinformatics 2012, 28, 373–380. [Google Scholar] [CrossRef] [Green Version]
- Monteleone, P.; Monteleone, A.M.; Troisi, J.; Dalle Grave, R.; Corrivetti, G.; Calugi, S.; Scala, G.; Patriciello, G.; Zanetti, A.; Maj, M. Metabolomics Signatures of Acutely Ill and Short-Term Weight Recovered Women with Anorexia Nervosa. Mol. Psychiatry 2019. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, M. An Introduction to Genetic Algorithms; MIT Press: Cambridge, MA, USA, 1998. [Google Scholar]
- Ward, J.H., Jr. Hierarchical Grouping to Optimize an Objective Function. J. Am. Stat. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
- Fluss, R.; Faraggi, D.; Reiser, B. Estimation of the Youden Index and Its Associated Cutoff Point. Biom. J. 2005, 47, 458–472. [Google Scholar] [CrossRef] [Green Version]
- Hsu, P.P.; Sabatini, D.M. Cancer Cell Metabolism: Warburg and Beyond. Cell 2008, 134, 703–707. [Google Scholar] [CrossRef] [Green Version]
- Warburg, O.H. Über Den Stoffwechsel Der Tumoren: Arbeiten Aus Dem Kaiser Wilhelm-Institut Für Biologie, Berlin-Dahlem; Kaiser-Wilhelm-Institut für Biologie/Julius Springer: Berlin, Germany, 1926. [Google Scholar]
- Coller, H.A. Is Cancer a Metabolic Disease? Am. J. Pathol. 2014, 184, 4–17. [Google Scholar] [CrossRef] [Green Version]
- Massari, F.; Ciccarese, C.; Santoni, M.; Iacovelli, R.; Mazzucchelli, R.; Piva, F.; Scarpelli, M.; Berardi, R.; Tortora, G.; Lopez-Beltran, A.; et al. Metabolic Phenotype of Bladder Cancer. Cancer Treat. Rev. 2016, 45, 46–57. [Google Scholar] [CrossRef]
- Afonso, J.; Santos, L.L.; Longatto-Filho, A.; Baltazar, F. Competitive Glucose Metabolism as a Target to Boost Bladder Cancer Immunotherapy. Nat. Rev. Urol. 2020, 17, 77–106. [Google Scholar] [CrossRef]
- Altenberg, B.; Greulich, K.O. Genes of Glycolysis Are Ubiquitously Overexpressed in 24 Cancer Classes. Genomics 2004, 84, 1014–1020. [Google Scholar] [CrossRef]
- Medina, R.A.; Owen, G.I. Glucose Transporters: Expression, Regulation and Cancer. Biol. Res. 2002, 35, 9–26. [Google Scholar] [CrossRef] [PubMed]
- Marín-Hernández, A.; Rodríguez-Enríquez, S.; Vital-González, P.A.; Flores-Rodríguez, F.L.; Macías-Silva, M.; Sosa-Garrocho, M.; Moreno-Sánchez, R. Determining and Understanding the Control of Glycolysis in Fast-Growth Tumor Cells. FEBS J. 2006, 273, 1975–1988. [Google Scholar] [CrossRef]
- Vora, S.; Halper, J.P.; Knowles, D.M. Alterations in the Activity and Isozymic Profile off Human Phosphofructokinase during Malignant Transformation in Vivo and in Vitro: Transformation- and Progression-Linked Discriminants off Malignancy. Cancer Res. 1985, 45, 2993–3001. [Google Scholar]
- Wu, M.; Lin, P.; Xu, L.; Yu, Z.; Chen, Q.; Gu, H.; Liu, C. Prognostic Role of Serum Lactate Dehydrogenase in Patients With Urothelial Carcinoma: A Systematic Review and Meta-Analysis. Front. Oncol. 2020, 10, 677. [Google Scholar] [CrossRef]
- Mendler, A.N.; Hu, B.; Prinz, P.U.; Kreutz, M.; Gottfried, E.; Noessner, E. Tumor Lactic Acidosis Suppresses CTL Function by Inhibition of P38 and JNK/c-Jun Activation. Int. J. Cancer 2012, 131, 633–640. [Google Scholar] [CrossRef]
- Fischer, K.; Hoffmann, P.; Voelkl, S.; Meidenbauer, N.; Ammer, J.; Edinger, M.; Gottfried, E.; Schwarz, S.; Rothe, G.; Hoves, S.; et al. Inhibitory Effect of Tumor Cell-Derived Lactic Acid on Human T Cells. Blood 2007, 109, 3812–3819. [Google Scholar] [CrossRef] [PubMed]
- Végran, F.; Boidot, R.; Michiels, C.; Sonveaux, P.; Feron, O. Lactate Influx through the Endothelial Cell Monocarboxylate Transporter MCT1 Supports an NF-ΚB/IL-8 Pathway That Drives Tumor Angiogenesis. Cancer Res. 2011, 71, 2550–2560. [Google Scholar] [CrossRef] [Green Version]
- Wise, D.R.; Thompson, C.B. Glutamine Addiction: A New Therapeutic Target in Cancer. Trends Biochem. Sci. 2010, 35, 427–433. [Google Scholar] [CrossRef] [Green Version]
- DeBerardinis, R.J.; Thompson, C.B. Cellular Metabolism and Disease: What Do Metabolic Outliers Teach Us? Cell 2012, 148, 1132–1144. [Google Scholar] [CrossRef] [Green Version]
- Amara, C.S.; Vantaku, V.; Lotan, Y.; Putluri, N. Recent Advances in the Metabolomic Study of Bladder Cancer. Expert Rev. Proteom. 2019, 16, 315–324. [Google Scholar] [CrossRef]
- Zhou, Y.; Song, R.; Zhang, Z.; Lu, X.; Zeng, Z.; Hu, C.; Liu, X.; Li, Y.; Hou, J.; Sun, Y.; et al. The Development of Plasma Pseudotargeted GC-MS Metabolic Profiling and Its Application in Bladder Cancer. Anal. Bioanal. Chem. 2016, 408, 6741–6749. [Google Scholar] [CrossRef]
- Loras, A.; Trassierra, M.; Sanjuan-Herráez, D.; Martínez-Bisbal, M.C.; Castell, J.V.; Quintás, G.; Ruiz-Cerdá, J.L. Bladder Cancer Recurrence Surveillance by Urine Metabolomics Analysis. Sci. Rep. 2018, 8, 9172. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Song, R.; Ma, C.; Zhou, L.; Liu, X.; Yin, P.; Zhang, Z.; Sun, Y.; Xu, C.; Lu, X.; et al. Discovery and Validation of Potential Urinary Biomarkers for Bladder Cancer Diagnosis Using a Pseudotargeted GC-MS Metabolomics Method. Oncotarget 2017, 8, 20719–20728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kouznetsova, V.L.; Kim, E.; Romm, E.L.; Zhu, A.; Tsigelny, I.F. Recognition of Early and Late Stages of Bladder Cancer Using Metabolites and Machine Learning. Metabolomics 2019, 15, 94. [Google Scholar] [CrossRef]
- Shao, C.-H.; Chen, C.-L.; Lin, J.-Y.; Chen, C.-J.; Fu, S.-H.; Chen, Y.-T.; Chang, Y.-S.; Yu, J.-S.; Tsui, K.-H.; Juo, C.-G.; et al. Metabolite Marker Discovery for the Detection of Bladder Cancer by Comparative Metabolomics. Oncotarget 2017, 8, 38802–38810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Den Besten, G.; van Eunen, K.; Groen, A.K.; Venema, K.; Reijngoud, D.-J.; Bakker, B.M. The Role of Short-Chain Fatty Acids in the Interplay between Diet, Gut Microbiota, and Host Energy Metabolism. J. Lipid Res. 2013, 54, 2325–2340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gunjur, A. Cancer and the Microbiome. Lancet Oncol. 2020, 21, 888. [Google Scholar] [CrossRef]
- Oresta, B.; Hurle, R.; Lazzeri, M.; Frego, N.; Saita, A.; Faccani, C.; Fasulo, V.; Casale, P.; Pozzi, C.; Guazzoni, G.F.; et al. Characterization of the Urinary Microbiota in Bladder Cancer Patients. J. Clin. Oncol. 2020, 38, 535. [Google Scholar] [CrossRef]
- Cimadamore, A.; Santoni, M.; Massari, F.; Gasparrini, S.; Cheng, L.; Lopez-Beltran, A.; Montironi, R.; Scarpelli, M. Microbiome and Cancers, With Focus on Genitourinary Tumors. Front. Oncol. 2019, 9, 178. [Google Scholar] [CrossRef]
- Bučević Popović, V.; Šitum, M.; Chow, C.-E.T.; Chan, L.S.; Roje, B.; Terzić, J. The Urinary Microbiome Associated with Bladder Cancer. Sci. Rep. 2018, 8, 12157. [Google Scholar] [CrossRef] [Green Version]
- Soini, J.; Falschlehner, C.; Liedert, C.; Bernhardt, J.; Vuoristo, J.; Neubauer, P. Norvaline Is Accumulated after a Down-Shift of Oxygen in Escherichia Coli W3110. Microb. Cell Factories 2008, 7, 30. [Google Scholar] [CrossRef] [Green Version]
- DeGeorge, K.C.; Holt, H.R.; Hodges, S.C. Bladder Cancer: Diagnosis and Treatment. Am. Fam. Physician 2017, 96, 507–514. [Google Scholar]
- Witjes, J.A.; Compérat, E.; Cowan, N.C.; De Santis, M.; Gakis, G.; Lebret, T.; Ribal, M.J.; Van der Heijden, A.G.; Sherif, A. EAU Guidelines on Muscle-Invasive and Metastatic Bladder Cancer: Summary of the 2013 Guidelines. Eur. Urol. 2014, 65, 778–792. [Google Scholar] [CrossRef] [PubMed]
- Onan, A. On the Performance of Ensemble Learning for Automated Diagnosis of Breast Cancer. In Artificial Intelligence Perspectives and Applications, Proceedings of the 4th Computer Science On-line Conference 2015 (CSOC2015), Online, 27–30 April 2015; Silhavy, R., Senkerik, R., Oplatkova, Z.K., Prokopova, Z., Silhavy, P., Eds.; Springer: Cham, Switzerland, 2015; pp. 119–129. [Google Scholar]
- Elrahman, S.M.A.; Abraham, A. A Review of Class Imbalance Problem. J. Netw. Innov. Comput. 2013, 1, 332–340. [Google Scholar]
- Wilk, T.; Wozniak, M. Complexity and Multithreaded Implementation Analysis of One Class-Classifiers Fuzzy Combiner. In Proceedings of the 6th International Conference on Hybrid Artificial Intelligent Systems—Volume Part II, Wroclaw, Poland, 23–25 May 2011; pp. 237–244. [Google Scholar]




| Low Grade Bladder Cancer (BCL) (n = 55) | High Grade Bladder Cancer (BCH) (n = 94) | Control (n = 81) | |
|---|---|---|---|
| Age (years) 1 | 68.4 (64.0–73.0)/46–87 | 73.3 (69.0–79.0)/49–88 | 67.3 (58.0–73.0)/47–87 |
| Men | 46 (83.6%) | 81 (86.2%) | 67 (82.7%) |
| Women | 9 (16.4%) | 13 (13.8%) | 14 (17.3%) |
| Non muscle invasive cancer (NMIC) | 55 (100%) (46 pTa; 9 pT1) 3 | 69 (73.4%) § (27 pTa; 42 pT1) | NA 2 |
| Muscle invasive cancer (MIC) | 0 | 26 (26.6%) | NA |
| Smokers | 33 (60.0%) * | 56 (59.6%) * | 20 (24.7%) |
| No-Smokers | 22 (40.0%) * | 38 (40.4) * | 61 (75.3%) |
| S 1 | Sp | PPV | NPV | PLR | NLR | A | ||
|---|---|---|---|---|---|---|---|---|
| Model I | Decision Tree | 0.97 ± 0.01 | 0.96 ± 0.02 | 0.98 ± 0.01 | 0.95 ± 0.04 | 26.28 | 0.03 | 0.97 |
| PLS-DA | 1.00 ± 0.00 | 0.96 ± 0.02 | 0.98 ± 0.01 | 1.00 ± 0.00 | 27.00 | 0.00 | 0.99 | |
| Naïve Bayes | 0.81 ± 0.03 | 0.99 ± 0.01 | 0.99 ± 0.01 | 0.74 ± 0.04 | 65.78 | 0.19 | 0.87 | |
| Random Forest | 0.97 ± 0.01 | 0.98 ± 0.02 | 0.99 ± 0.01 | 0.95 ± 0.02 | 39.41 | 0.03 | 0.97 | |
| k-NN | 1.00 ± 0.00 | 1.00 ± 0.00 | 1.00 ± 0.00 | 1.00 ± 0.00 | ND | 0.00 | 1.00 | |
| aNN | 1.00 ± 0.00 | 0.99 ± 0.01 | 0.99 ± 0.01 | 1.00 ± 0.01 | 81.00 | 0.00 | 1.00 | |
| Logistic Regression | 1.00 ± 0.00 | 1.00 ± 0.00 | 1.00 ± 0.00 | 1.00 ± 0.00 | ND | 0.00 | 1.00 | |
| SVM | 1.00 ± 0.00 | 0.98 ± 0.02 | 0.99 ± 0.01 | 1.00 ± 0.00 | 40.50 | 0.00 | 0.99 | |
| Deep Learning | 1.00 ± 0.00 | 0.99 ± 0.01 | 0.99 ± 0.01 | 1.00 ± 0.01 | 81.00 | 0.00 | 1.00 | |
| Ensemble | 1.00 ± 0.00 | 0.98 ± 0.02 | 0.99 ± 0.01 | 1.00 ± 0.00 | 40.50 | 0.00 | 0.99 | |
| Model II | Decision Tree | 0.94 ± 0.03 | 0.85 ± 0.05 | 0.92 ± 0.03 | 0.89 ± 0.04 | 6.44 | 0.07 | 0.91 |
| PLS-DA | 0.64 ± 0.05 | 0.89 ± 0.04 | 0.91 ± 0.04 | 0.60 ± 0.05 | 5.96 | 0.41 | 0.73 | |
| Naïve Bayes | 0.46 ± 0.05 | 0.93 ± 0.04 | 0.91 ± 0.04 | 0.50 ± 0.05 | 6.29 | 0.59 | 0.63 | |
| Random Forest | 0.98 ± 0.01 | 0.56 ± 0.07 | 0.79 ± 0.04 | 0.94 ± 0.04 | 2.24 | 0.04 | 0.83 | |
| k-NN | 1.00 ± 0.00 | 1.00 ± 0.00 | 1.00 ± 0.00 | 1.00 ± 0.00 | ND | 0.00 | 1.00 | |
| aNN | 0.99 ± 0.01 | 0.35 ± 0.06 | 0.72 ± 0.04 | 0.95 ± 0.05 | 1.51 | 0.03 | 0.75 | |
| Logistic Regression | 1.00 ± 0.00 | 1.00 ± 0.00 | 1.00 ± 0.00 | 1.00 ± 0.00 | ND | 0.00 | 1.00 | |
| SVM | 0.98 ± 0.01 | 0.27 ± 0.06 | 0.70 ± 0.04 | 0.88 ± 0.08 | 1.35 | 0.08 | 0.72 | |
| Deep Learning | 0.86 ± 0.04 | 0.80 ± 0.05 | 0.88 ± 0.03 | 0.77 ± 0.06 | 4.31 | 0.17 | 0.84 | |
| Ensemble | 0.79 ± 0.06 | 0.83 ± 0.07 | 0.87 ± 0.05 | 0.73 ± 0.08 | 4.56 | 0.26 | 0.80 | |
| Model III | Decision Tree | 1.00 ± 0.00 | 0.20 ± 0.13 | 0.93 ± 0.03 | 1.00 ± 0.00 | 1.25 | 0.00 | 0.94 |
| PLS-DA | 0.51 ± 0.05 | 0.50 ± 0.11 | 0.86 ± 0.04 | 0.15 ± 0.04 | 1.03 | 0.97 | 0.51 | |
| Naïve Bayes | 0.49 ± 0.05 | 0.90 ± 0.07 | 0.97 ± 0.02 | 0.24 ± 0.05 | 4.91 | 0.57 | 0.55 | |
| Random Forest | 1.00 ± 0.00 | 0.00 ± 0.00 | 0.85 ± 0.03 | ND | 1.00 | ND | 0.85 | |
| k-NN | 1.00 ± 0.00 | 1.00 ± 0.00 | 1.00 ± 0.00 | 1.00 ± 0.00 | ND | 0.00 | 1.00 | |
| aNN | 1.00 ± 0.00 | 0.30 ± 0.10 | 0.89 ± 0.03 | 1.00 ± 0.00 | 1.43 | 0.00 | 0.90 | |
| Logistic Regression | 1.00 ± 0.00 | 1.00 ± 0.00 | 1.00 ± 0.00 | 1.00 ± 0.00 | ND | 0.00 | 1.00 | |
| SVM | 1.00 ± 0.00 | 0.10 ± 0.07 | 0.86 ± 0.03 | 1.00 ± 0.00 | 1.11 | 0.00 | 0.87 | |
| Deep Learning | 1.00 ± 0.00 | 0.50 ± 0.11 | 0.92 ± 0.02 | 1.00 ± 0.00 | 2.00 | 0.00 | 0.93 | |
| Ensemble | 1.00 ± 0.00 | 0.00 ± 0.00 | 0.85 ± 0.03 | ND | 1.00 | ND | 0.85 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Troisi, J.; Colucci, A.; Cavallo, P.; Richards, S.; Symes, S.; Landolfi, A.; Scala, G.; Maiorino, F.; Califano, A.; Fabiano, M.; et al. A Serum Metabolomic Signature for the Detection and Grading of Bladder Cancer. Appl. Sci. 2021, 11, 2835. https://doi.org/10.3390/app11062835
Troisi J, Colucci A, Cavallo P, Richards S, Symes S, Landolfi A, Scala G, Maiorino F, Califano A, Fabiano M, et al. A Serum Metabolomic Signature for the Detection and Grading of Bladder Cancer. Applied Sciences. 2021; 11(6):2835. https://doi.org/10.3390/app11062835
Chicago/Turabian StyleTroisi, Jacopo, Angelo Colucci, Pierpaolo Cavallo, Sean Richards, Steven Symes, Annamaria Landolfi, Giovanni Scala, Francesco Maiorino, Alfonso Califano, Marco Fabiano, and et al. 2021. "A Serum Metabolomic Signature for the Detection and Grading of Bladder Cancer" Applied Sciences 11, no. 6: 2835. https://doi.org/10.3390/app11062835
APA StyleTroisi, J., Colucci, A., Cavallo, P., Richards, S., Symes, S., Landolfi, A., Scala, G., Maiorino, F., Califano, A., Fabiano, M., Silvestre, G., Mastella, F., Caputo, A., D’Antonio, A., & Altieri, V. (2021). A Serum Metabolomic Signature for the Detection and Grading of Bladder Cancer. Applied Sciences, 11(6), 2835. https://doi.org/10.3390/app11062835







