A Comparative Cross-Platform Meta-Analysis to Identify Potential Biomarker Genes Common to Endometriosis and Recurrent Pregnancy Loss
Abstract
1. Introduction
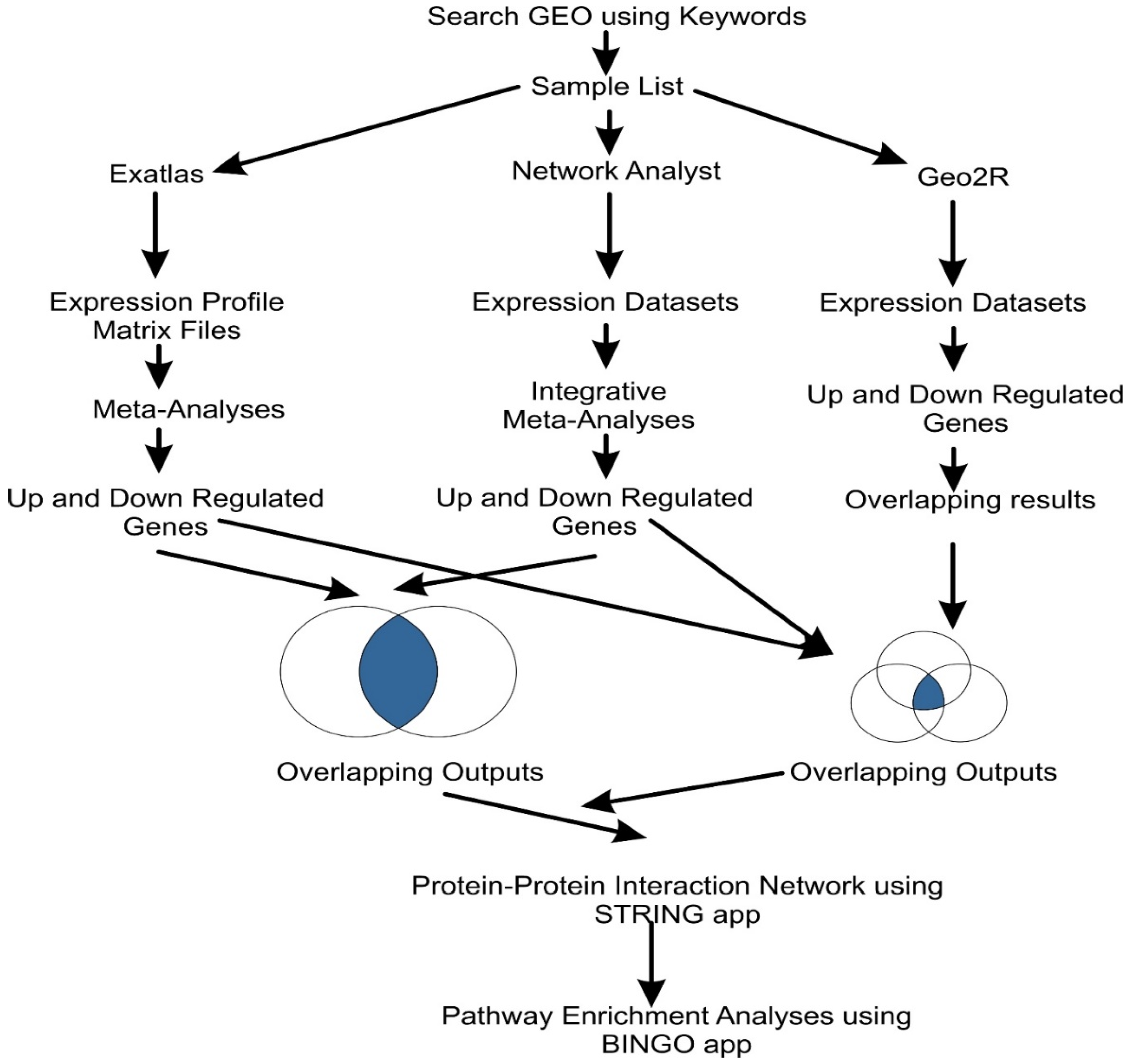
2. Materials and Methods
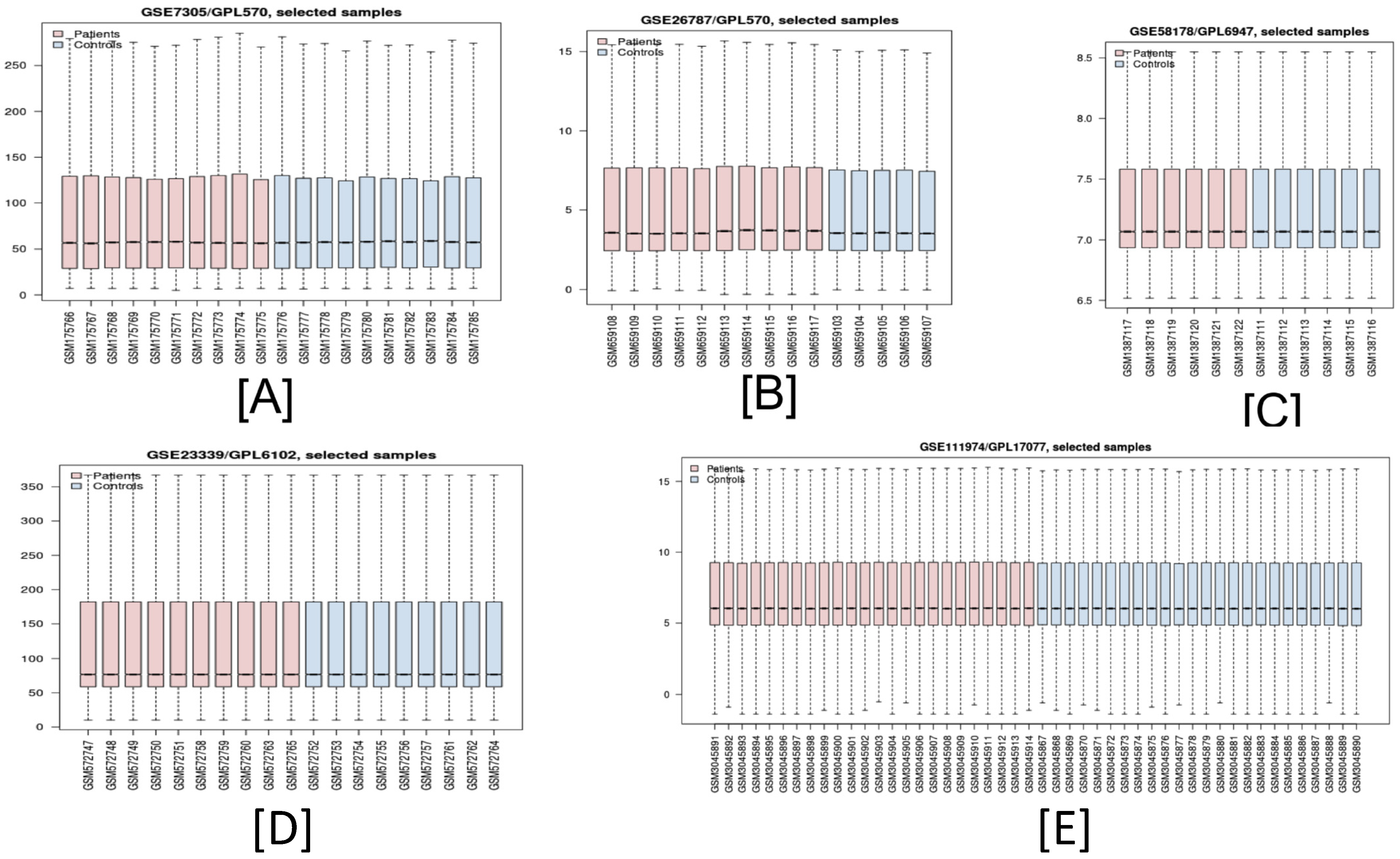
2.1. Microarray Data
2.2. DEG Screening and Meta-Analyses
2.3. Comparative Analyses
2.4. Protein–Protein Interaction Network Construction and Pathway Enrichment Analyses
2.4.1. Protein–Protein Network Interaction
2.4.2. Pathway Enrichment Analysis
3. Results
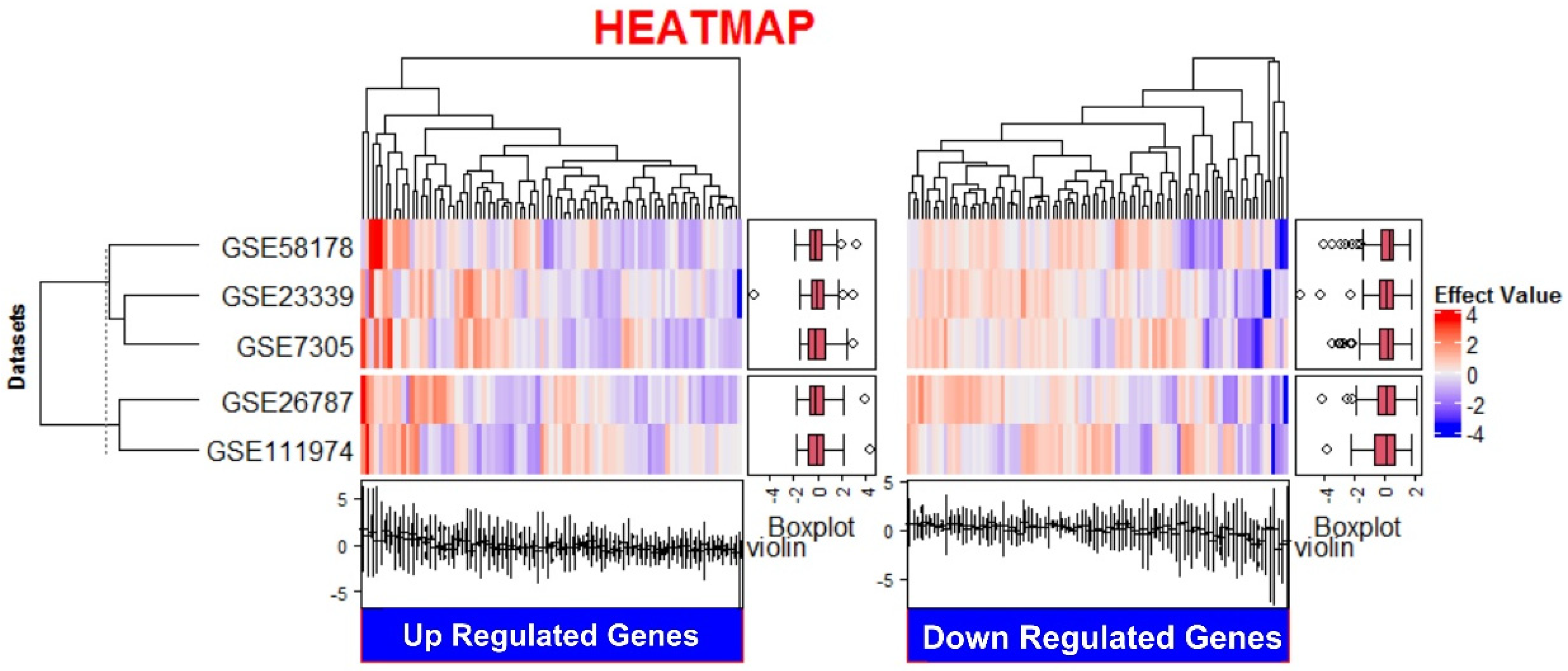
3.1. Expression of Up- and Down-Regulated Genes
3.2. Protein–Protein Interaction (PPI) Network
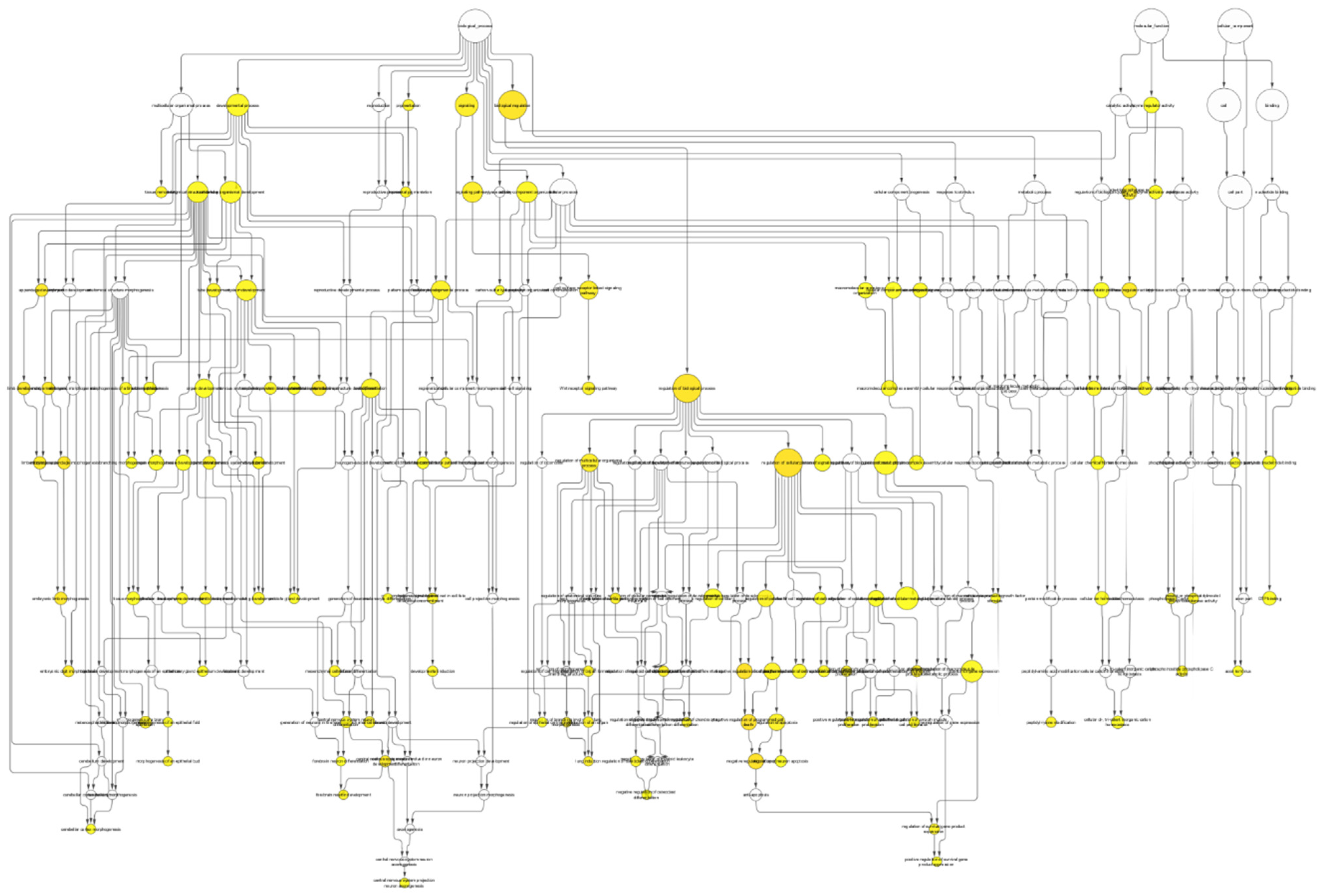
3.3. Pathway Enrichment Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Farquhar, C. Endometriosis. BMJ 2007, 334, 249–253. [Google Scholar] [CrossRef]
- Klemmt, P.A.B.; Starzinski-Powitz, A. Molecular and Cellular Pathogenesis of Endometriosis. Curr. Womens Health Rev. 2018, 14, 106–116. [Google Scholar] [CrossRef]
- Laganà, A.S.; Garzon, S.; Götte, M.; Viganò, P.; Franchi, M.; Ghezzi, F.; Martin, D.C. The Pathogenesis of Endometriosis: Molecular and Cell Biology Insights. Int. J. Mol. Sci. 2019, 20, 5615. [Google Scholar] [CrossRef]
- Pazhohan, A.; Amidi, F.; Akbari-Asbagh, F.; Seyedrezazadeh, E.; Farzadi, L.; Khodarahmin, M.; Mehdinejadiani, S.; Sobhani, A. The Wnt/β-catenin signaling in endometriosis, the expression of total and active forms of β-catenin, total and inactive forms of glycogen synthase kinase-3β, WNT7a and DICKKOPF-1. Eur. J. Obstet. Gynecol. Reprod. Biol. 2018, 220, 1–5. [Google Scholar] [CrossRef]
- RPL (Recurrent Pregnancy Loss): Guideline of the European Society of Human Reproduction and Embryology. ESHRE Early Pregnancy Guideline Development Group. 2017, pp. 1–153. Available online: https://www.eshre.eu/Guidelines-and-Legal/Guidelines/Recurrent-pregnancy-loss.aspx (accessed on 18 July 2020).
- Imanaka, S.; Maruyama, S.; Kimura, M.; Nagayasu, M.; Kobayashi, H. Towards an understanding of the molecular mechanisms of endometriosis-associated symptoms (Review). World Acad. Sci. J. 2020, 2, 12. [Google Scholar] [CrossRef]
- Ticconi, C.; Pietropolli, A.; Di Simone, N.; Piccione, E.; Fazleabas, A. Endometrial Immune Dysfunction in Recurrent Pregnancy Loss. Int. J. Mol. Sci. 2019, 20, 5332. [Google Scholar] [CrossRef] [PubMed]
- Sapkota, Y.; iPSYCH-SSI-Broad Group; Steinthorsdottir, V.; Morris, A.P.; Fassbender, A.; Rahmioglu, N.; De Vivo, I.; Buring, J.E.; Zhang, F.; Edwards, T.L.; et al. Meta-analysis identifies five novel loci associated with endometriosis highlighting key genes involved in hormone metabolism. Nat. Commun. 2017, 8, 15539. [Google Scholar] [CrossRef]
- Akter, S.; Xu, D.; Nagel, S.C.; Bromfield, J.J.; Pelch, K.; Wilshire, G.B.; Joshi, T. Machine Learning Classifiers for Endometriosis Using Transcriptomics and Methylomics Data. Front. Genet. 2019, 10, 766. [Google Scholar] [CrossRef]
- Méar, L.; Herr, M.; Fauconnier, A.; Pineau, C.; Vialard, F. Polymorphisms and endometriosis: A systematic review and meta-analyses. Hum. Reprod. Update 2020, 26, 73–102. [Google Scholar] [CrossRef]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Sharov, A.A.; Schlessinger, D.; Ko, M.S. ExAtlas: An interactive online tool for meta-analysis of gene expression data. J. Bioinform. Comput. Biol. 2015, 13, 1550019. [Google Scholar] [CrossRef]
- Zhou, G.; Soufan, O.; Ewald, J.; Hancock, R.E.W.; Basu, N.; Xia, J. NetworkAnalyst 3.0: A visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019, 47, W234–W241. [Google Scholar] [CrossRef]
- Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Available online: https://pubmed.ncbi.nlm.nih.gov/25605792/ (accessed on 23 March 2021).
- Mudunuri, U.; Che, A.; Yi, M.; Stephens, R.M. bioDBnet: The biological database network. Bioinformatics 2009, 25, 555–556. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Eils, R.; Schlesner, M. Complex Heatmaps Reveal Patterns and Correlations in Multidimensional Genomic Data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Doncheva, N.T.; Morris, J.H.; Gorodkin, J.; Jensen, L.J. Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J. Proteome Res. 2019, 18, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Maere, S.; Heymans, K.; Kuiper, M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005, 21, 3448–3449. [Google Scholar] [CrossRef] [PubMed]
- Monsivais, D.; Dyson, M.T.; Yin, P.; Coon, J.S.; Navarro, A.; Feng, G.; Malpani, S.S.; Ono, M.; Ercan, C.M.; Wei, J.J.; et al. ERbeta- and prostaglandin E2-regulated pathways integrate cell proliferation via Ras-like and estrogen-regulated growth inhibitor in endometriosis. Mol. Endocrinol. 2014, 28, 1304–1315. [Google Scholar] [CrossRef]
- Hawkins, S.M.; Creighton, C.J.; Han, D.Y.; Zariff, A.; Anderson, M.L.; Gunaratne, P.H.; Matzuk, M.M. Functional microRNA involved in endometriosis. Mol. Endocrinol. 2011, 25, 821–832. [Google Scholar] [CrossRef]
- Hever, A.; Roth, R.B.; Hevezi, P.; Marin, M.E.; Acosta, J.A.; Acosta, H.; Rojas, J.; Herrera, R.; Grigoriadis, D.; White, E.; et al. Human endometriosis is associated with plasma cells and overexpression of B lymphocyte stimulator. Proc. Natl. Acad. Sci. USA 2007, 104, 12451–12456. [Google Scholar] [CrossRef]
- Bastu, E.; Demiral, I.; Gunel, T.; Ulgen, E.; Gumusoglu, E.; Hosseini, M.K.; Sezerman, U.; Buyru, F.; Yeh, J. Potential Marker Pathways in the Endometrium That May Cause Recurrent Implantation Failure. Reprod. Sci. 2019, 26, 879–890. [Google Scholar] [CrossRef]
- Ledee, N.; Munaut, C.; Aubert, J.; Serazin, V.; Rahmati, M.; Chaouat, G.; Sandra, O.; Foidart, J.M. Specific and extensive endometrial deregulation is present before conception in IVF/ICSI repeated implantation failures (IF) or recurrent miscarriages. J. Pathol. 2011, 225, 554–564. [Google Scholar] [CrossRef] [PubMed]
- Hyde, K.J.; Schust, D.J. Genetic considerations in recurrent pregnancy loss. Cold Spring Harb. Perspect. Med. 2015, 5, a023119. [Google Scholar] [CrossRef] [PubMed]
- Kacprzak, M.; Chrzanowska, M.; Skoczylas, B.; Moczulska, H.; Borowiec, M.; Sieroszewski, P. Genetic causes of recurrent miscarriages. Ginekol. Pol. 2016, 87, 722–726. [Google Scholar] [CrossRef]
- Kaser, D. The Status of Genetic Screening in Recurrent Pregnancy Loss. Obstet. Gynecol. Clin. N. Am. 2018, 45, 143–154. [Google Scholar] [CrossRef]
- Moghbeli, M. Genetics of recurrent pregnancy loss among Iranian population. Mol. Genet. Genom. Med. 2019, 7, e891. [Google Scholar] [CrossRef]
- Vaiman, D. Genetic regulation of recurrent spontaneous abortion in humans. Biomed. J. 2015, 38, 11–24. [Google Scholar] [CrossRef]
- Hansen, K.A.; Eyster, K.M. Genetics and genomics of endometriosis. Clin. Obstet. Gynecol. 2010, 53, 403–412. [Google Scholar] [CrossRef]
- Bischoff, F.; Simpson, J.L. Genetics of endometriosis: Heritability and candidate genes. Best Pract. Res. Clin. Obstet. Gynaecol. 2004, 18, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Vassilopoulou, L.; Matalliotakis, M.; Zervou, M.I.; Matalliotaki, C.; Krithinakis, K.; Matalliotakis, I.; Spandidos, D.A.; Goulielmos, G.N. Defining the genetic profile of endometriosis. Exp. Ther. Med. 2019, 17, 3267–3281. [Google Scholar] [CrossRef]
- Rahmioglu, N.; Montgomery, G.W.; Zondervan, K.T. Genetics of endometriosis. Womens Health 2015, 11, 577–586. [Google Scholar] [CrossRef]
- Tomassetti, C.; Meuleman, C.; Pexsters, A.; Mihalyi, A.; Kyama, C.; Simsa, P.; D’Hooghe, T.M. Endometriosis, recurrent miscarriage and implantation failure: Is there an immunological link? Reprod. Biomed. Online 2006, 13, 58–64. [Google Scholar] [CrossRef]
- Santulli, P.; Marcellin, L.; Menard, S.; Thubert, T.; Khoshnood, B.; Gayet, V.; Goffinet, F.; Ancel, P.Y.; Chapron, C. Increased rate of spontaneous miscarriages in endometriosis-affected women. Hum. Reprod. 2016, 31, 1014–1023. [Google Scholar] [CrossRef]
- Kohl Schwartz, A.S.; Wolfler, M.M.; Mitter, V.; Rauchfuss, M.; Haeberlin, F.; Eberhard, M.; von Orelli, S.; Imthurn, B.; Imesch, P.; Fink, D.; et al. Endometriosis, especially mild disease: A risk factor for miscarriages. Fertil. Steril. 2017, 108, 806–814.e802. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Wang, Y.; Wu, Z.; Pan, N.; Yan, L.; Ma, C. Risk of miscarriage in women with endometriosis undergoing IVF fresh cycles: A retrospective cohort study. Reprod. Biol. Endocrinol. 2019, 17, 21. [Google Scholar] [CrossRef]
- Poli-Neto, O.B.; Meola, J.; Rosa-E-Silva, J.C.; Tiezzi, D. Transcriptome meta-analysis reveals differences of immune profile between eutopic endometrium from stage I-II and III-IV endometriosis independently of hormonal milieu. Sci. Rep. 2020, 10, 1–17. [Google Scholar] [CrossRef]
- Gou, J.; Hu, T.; Li, L.; Xue, L.; Zhao, X.; Yi, T.; Li, Z. Role of epithelial-mesenchymal transition regulated by twist basic helix-loop-helix transcription factor 2 (Twist2) in embryo implantation in mice. Reprod. Fertil. Dev. 2019, 31, 932–940. [Google Scholar] [CrossRef] [PubMed]
- Girisha, K.M.; Bidchol, A.M.; Sarpangala, M.K.; Satyamoorthy, K. A novel frameshift mutation in TWIST2 gene causing Setleis syndrome. Indian J. Pediatr. 2014, 81, 302–304. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Meng, T.; Wang, S.; Zhang, H.; Mues, G.; Qin, C.; Feng, J.Q.; D’Souza, R.N.; Lu, Y. Twist1- and Twist2-haploinsufficiency results in reduced bone formation. PLoS ONE 2014, 9, e99331. [Google Scholar] [CrossRef]
- Franco, H.L.; Casasnovas, J.; Rodriguez-Medina, J.R.; Cadilla, C.L. Redundant or separate entities?—Roles of Twist1 and Twist2 as molecular switches during gene transcription. Nucleic Acids Res. 2011, 39, 1177–1186. [Google Scholar] [CrossRef]
- Gong, X.Q.; Li, L. Dermo-1, a multifunctional basic helix-loop-helix protein, represses MyoD transactivation via the HLH domain, MEF2 interaction, and chromatin deacetylation. J. Biol. Chem. 2002, 277, 12310–12317. [Google Scholar] [CrossRef]
- Spicer, D.B.; Rhee, J.; Cheung, W.L.; Lassar, A.B. Inhibition of myogenic bHLH and MEF2 transcription factors by the bHLH protein Twist. Science 1996, 272, 1476–1480. [Google Scholar] [CrossRef]
- Lee, M.S.; Lowe, G.; Flanagan, S.; Kuchler, K.; Glackin, C.A. Human Dermo-1 has attributes similar to twist in early bone development. Bone 2000, 27, 591–602. [Google Scholar] [CrossRef]
- Bialek, P.; Kern, B.; Yang, X.; Schrock, M.; Sosic, D.; Hong, N.; Wu, H.; Yu, K.; Ornitz, D.M.; Olson, E.N.; et al. A twist code determines the onset of osteoblast differentiation. Dev. Cell 2004, 6, 423–435. [Google Scholar] [CrossRef]
- Sharabi, A.B.; Aldrich, M.; Sosic, D.; Olson, E.N.; Friedman, A.D.; Lee, S.H.; Chen, S.Y. Twist-2 controls myeloid lineage development and function. PLoS Biol. 2008, 6, e316. [Google Scholar] [CrossRef] [PubMed]
- Kallio, H.; Pastorekova, S.; Pastorek, J.; Waheed, A.; Sly, W.S.; Mannisto, S.; Heikinheimo, M.; Parkkila, S. Expression of carbonic anhydrases IX and XII during mouse embryonic development. BMC Dev. Biol. 2006, 6, 22. [Google Scholar] [CrossRef] [PubMed]
- Agafonov, D.E.; Kastner, B.; Dybkov, O.; Hofele, R.V.; Liu, W.T.; Urlaub, H.; Luhrmann, R.; Stark, H. Molecular architecture of the human U4/U6.U5 tri-snRNP. Science 2016, 351, 1416–1420. [Google Scholar] [CrossRef] [PubMed]
- Chari, A.; Golas, M.M.; Klingenhager, M.; Neuenkirchen, N.; Sander, B.; Englbrecht, C.; Sickmann, A.; Stark, H.; Fischer, U. An assembly chaperone collaborates with the SMN complex to generate spliceosomal SnRNPs. Cell 2008, 135, 497–509. [Google Scholar] [CrossRef] [PubMed]
- Grimm, C.; Chari, A.; Pelz, J.P.; Kuper, J.; Kisker, C.; Diederichs, K.; Stark, H.; Schindelin, H.; Fischer, U. Structural basis of assembly chaperone-mediated snRNP formation. Mol. Cell 2013, 49, 692–703. [Google Scholar] [CrossRef] [PubMed]
- Jurica, M.S.; Licklider, L.J.; Gygi, S.R.; Grigorieff, N.; Moore, M.J. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA 2002, 8, 426–439. [Google Scholar] [CrossRef]
- Kondo, Y.; Oubridge, C.; van Roon, A.M.; Nagai, K. Crystal structure of human U1 snRNP, a small nuclear ribonucleoprotein particle, reveals the mechanism of 5′ splice site recognition. Elife 2015, 4, e04986. [Google Scholar] [CrossRef] [PubMed]
- Pomeranz Krummel, D.A.; Oubridge, C.; Leung, A.K.; Li, J.; Nagai, K. Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature 2009, 458, 475–480. [Google Scholar] [CrossRef]
- Zhang, X.; Yan, C.; Hang, J.; Finci, L.I.; Lei, J.; Shi, Y. An Atomic Structure of the Human Spliceosome. Cell 2017, 169, 918–929.e14. [Google Scholar] [CrossRef] [PubMed]
- Bertram, K.; Agafonov, D.E.; Dybkov, O.; Haselbach, D.; Leelaram, M.N.; Will, C.L.; Urlaub, H.; Kastner, B.; Luhrmann, R.; Stark, H. Cryo-EM Structure of a Pre-catalytic Human Spliceosome Primed for Activation. Cell 2017, 170, 701–713.e11. [Google Scholar] [CrossRef] [PubMed]
- Bertram, K.; Agafonov, D.E.; Liu, W.T.; Dybkov, O.; Will, C.L.; Hartmuth, K.; Urlaub, H.; Kastner, B.; Stark, H.; Luhrmann, R. Cryo-EM structure of a human spliceosome activated for step 2 of splicing. Nature 2017, 542, 318–323. [Google Scholar] [CrossRef]
- Lillehoj, E.P.; Lu, W.; Kiser, T.; Goldblum, S.E.; Kim, K.C. MUC1 inhibits cell proliferation by a beta-catenin-dependent mechanism. Biochim. Biophys. Acta 2007, 1773, 1028–1038. [Google Scholar] [CrossRef] [PubMed]
- Weiske, J.; Albring, K.F.; Huber, O. The tumor suppressor Fhit acts as a repressor of beta-catenin transcriptional activity. Proc. Natl. Acad. Sci. USA 2007, 104, 20344–20349. [Google Scholar] [CrossRef]
- Bahmanyar, S.; Kaplan, D.D.; Deluca, J.G.; Giddings, T.H., Jr.; O’Toole, E.T.; Winey, M.; Salmon, E.D.; Casey, P.J.; Nelson, W.J.; Barth, A.I. beta-Catenin is a Nek2 substrate involved in centrosome separation. Genes Dev. 2008, 22, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ray, G.; Yoo, B.H.; Erdogan, M.; Rosen, K.V. Down-regulation of death-associated protein kinase-2 is required for beta-catenin-induced anoikis resistance of malignant epithelial cells. J. Biol. Chem. 2009, 284, 2012–2022. [Google Scholar] [CrossRef]
- Fiset, A.; Xu, E.; Bergeron, S.; Marette, A.; Pelletier, G.; Siminovitch, K.A.; Olivier, M.; Beauchemin, N.; Faure, R.L. Compartmentalized CDK2 is connected with SHP-1 and beta-catenin and regulates insulin internalization. Cell. Signal. 2011, 23, 911–919. [Google Scholar] [CrossRef]
- Satow, R.; Shitashige, M.; Jigami, T.; Fukami, K.; Honda, K.; Kitabayashi, I.; Yamada, T. beta-catenin inhibits promyelocytic leukemia protein tumor suppressor function in colorectal cancer cells. Gastroenterology 2012, 142, 572–581. [Google Scholar] [CrossRef]
- Genovese, G.; Ghosh, P.; Li, H.; Rettino, A.; Sioletic, S.; Cittadini, A.; Sgambato, A. The tumor suppressor HINT1 regulates MITF and beta-catenin transcriptional activity in melanoma cells. Cell Cycle 2012, 11, 2206–2215. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Wu, J.; Wang, Y.; Zhao, T.; Ma, B.; Liu, Y.; Fang, W.; Zhu, W.G.; Zhang, H. Kindlin 2 forms a transcriptional complex with beta-catenin and TCF4 to enhance Wnt signalling. EMBO Rep. 2012, 13, 750–758. [Google Scholar] [CrossRef]
- Bellows, T.S.; Fisher, T.W. Handbook of Biological Control: Principles and Applications of Biological Control; Academic Press: San Diego, CA, USA, 1999; p. xxiii. [Google Scholar]
- Brembeck, F.H.; Rosario, M.; Birchmeier, W. Balancing cell adhesion and Wnt signaling, the key role of beta-catenin. Curr. Opin. Genet. Dev. 2006, 16, 51–59. [Google Scholar] [CrossRef]
- Joksimovic, M.; Patel, M.; Taketo, M.M.; Johnson, R.; Awatramani, R. Ectopic Wnt/beta-catenin signaling induces neurogenesis in the spinal cord and hindbrain floor plate. PLoS ONE 2012, 7, e30266. [Google Scholar] [CrossRef]
- Kurnit, K.C.; Kim, G.N.; Fellman, B.M.; Urbauer, D.L.; Mills, G.B.; Zhang, W.; Broaddus, R.R. CTNNB1 (beta-catenin) mutation identifies low grade, early stage endometrial cancer patients at increased risk of recurrence. Mod. Pathol. 2017, 30, 1032–1041. [Google Scholar] [CrossRef]
- Yoh, K.; Prywes, R. Pathway Regulation of p63, a Director of Epithelial Cell Fate. Front. Endocrinol. 2015, 6, 51. [Google Scholar] [CrossRef] [PubMed]
- Fete, M.; vanBokhoven, H.; Clements, S.E.; McKeon, F.; Roop, D.R.; Koster, M.I.; Missero, C.; Attardi, L.D.; Lombillo, V.A.; Ratovitski, E.; et al. International Research Symposium on Ankyloblepharon-Ectodermal Defects-Cleft Lip/Palate (AEC) syndrome. Am. J. Med. Genet. A 2009, 149A, 1885–1893. [Google Scholar] [CrossRef]
- Fomenkov, A.; Huang, Y.P.; Topaloglu, O.; Brechman, A.; Osada, M.; Fomenkova, T.; Yuriditsky, E.; Trink, B.; Sidransky, D.; Ratovitski, E. P63 alpha mutations lead to aberrant splicing of keratinocyte growth factor receptor in the Hay-Wells syndrome. J. Biol. Chem. 2003, 278, 23906–23914. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Yang, M.; Guo, H.; Yang, L.; Wu, J.; Li, R.; Liu, P.; Lian, Y.; Zheng, X.; Yan, J.; et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 2013, 20, 1131–1139. [Google Scholar] [CrossRef] [PubMed]


| Sl. No. | GEO Accession | Subjec | Sample | Analytical Platform | Patient Type | Reference | ||
|---|---|---|---|---|---|---|---|---|
| Patient | Control | Total | ||||||
| 1 | GSE58178 | 6 | 6 | 12 | Endometrial tissue | GPL6947 (Illumina Human HT-12 v3.0 Expression Beadchip) | Endometriosis | [20] |
| 2 | GSE23339 | 10 | 9 | 19 | Endometrial tissue | GPL6102 (Illumina Human-6 v2.0 Expression Beadchip) | Endometriosis | [21] |
| 3 | GSE7305 | 10 | 10 | 20 | Endometrial tissue | GPL570 [HG-U133_Plus_2] (Affymetrix Human Genome U133 plus 2.0 Array) | Endometriosis | [22] |
| 4 | GSE111974 | 24 | 24 | 48 | Endometrial tissue | GPL17077 (Agilent-039494 SurePrint G3 Human GE v2 8 × 60K Microarray) | Recurrent Pregnancy Loss | [23] |
| 5 | GSE26787 | 10 | 5 | 15 | Endometrium | GPL570 [HG-U133_Plus_2] (Affymetrix Human Genome U133 Plus 2.0 Array) | Recurrent Pregnancy Loss | [24] |
| Gene Symbol | Entrez ID | Log Ratio Combined | Fold Change | FDR * |
|---|---|---|---|---|
| 1.1. TWIST2 | Twist Family Bhlh Transcription Factor 2 | −0.5434 | 3.494 | 8.79 × 10−11 |
| CA12 | Carbonic Anhydrase XII | −0.5111 | 3.244 | 0.001487 |
| PGBD5 | PiggyBac Transposable Element Derived 5 | −0.4782 | 3.007 | 0.002422 |
| H19 | H19, Imprinted Maternally Expressed Transcript (Non-Protein Coding) | −0.4696 | 2.948 | 0.000894 |
| SGCD | Sarcoglycan Delta | 0.4523 | 2.833 | 0 |
| ANO4 | Anoctamin 4 | −0.4227 | 2.647 | 2.42 × 10−5 |
| CHN2 | Chimerin 2 | 0.4002 | 2.513 | 8.01 × 10−7 |
| MLPH | Melanophilin | −0.3955 | 2.486 | 3.27 ×10−6 |
| PLPP1 | Phospholipid Phosphatase 1 | −0.3872 | 2.439 | 0.004665 |
| NR4A2 | Nuclear Receptor Subfamily 4 Group A Member 2 | 0.3829 | 2.415 | 0.0217 |
| DACH1 | Dachshund Family Transcription Factor 1 | −0.3827 | 2.414 | 3.07 ×10−8 |
| ADAMTS19 | ADAM Metallopeptidase With Thrombospondin Type 1 Motif 19 | −0.3787 | 2.392 | 0.004645 |
| VLDLR | Very Low-Density Lipoprotein Receptor | 0.3534 | 2.256 | 0.007674 |
| NFIB | Nuclear Factor I/B | 0.3519 | 2.249 | 4.80 × 10−6 |
| PCSK6 | Proprotein Convertase Subtilisin/Kexin Type 6 | 0.3468 | 2.223 | 0.0154 |
| GALNT10 | Polypeptide N-Acetylgalactosaminyltransferase 10 | 0.334 | 2.158 | 0 |
| TGM2 | Transglutaminase 2 | −0.3236 | 2.107 | 0.006722 |
| CREG1 | Cellular Repressor Of E1A-Stimulated Genes 1 | 0.3113 | 2.048 | 0.0175 |
| NDRG2 | NDRG Family Member 2 | 0.31 | 2.042 | 1.71 × 10−5 |
| H4C3 | H4 Clustered Histone 3 | −0.304 | 2.014 | 4.67 × 10−7 |
| RSPO3 | R-Spondin 3 | −0.3029 | 2.009 | 0.004831 |
| TSPAN2 | Tetraspanin 2 | 0.2999 | 1.995 | 0.0251 |
| CPXM1 | Carboxypeptidase X (M14 Family), Member 1 | −0.2865 | 1.934 | 4.13 × 10−6 |
| FBLN7 | Fibulin 7 | −0.2862 | 1.933 | 5.63 × 10−6 |
| HOXD11 | Homeobox D11 | −0.2822 | 1.915 | 0.0406 |
| Name | Average Shortest Path Length | Betweenness Centrality | Closeness Centrality | Clustering Coefficient | Degree |
|---|---|---|---|---|---|
| SNRPF | 2.145228 | 0.001357 | 0.466151 | 0.883247 | 84 |
| CTNNB1 | 1.929461 | 0.065249 | 0.51828 | 0.26485 | 54 |
| HNRNPAB | 2.373444 | 2.87E-05 | 0.421329 | 0.980408 | 50 |
| RBBP4 | 2.394191 | 0.002108 | 0.417678 | 0.642105 | 20 |
| WNT2 | 2.481328 | 0.000697 | 0.40301 | 0.760234 | 19 |
| PRKAB1 | 2.489627 | 0.003624 | 0.401667 | 0.79085 | 18 |
| GNAQ | 2.556017 | 0.009064 | 0.391234 | 0.333333 | 18 |
| GLI2 | 2.456432 | 0.002979 | 0.407095 | 0.698529 | 17 |
| RRAGD | 2.697095 | 0.000347 | 0.370769 | 0.95 | 16 |
| MITF | 2.448133 | 0.010795 | 0.408475 | 0.549451 | 14 |
| NES | 2.53112 | 0.000352 | 0.395082 | 0.769231 | 13 |
| TLE4 | 2.385892 | 0.000377 | 0.41913 | 0.709091 | 11 |
| RND3 | 2.53112 | 0.00226 | 0.395082 | 0.490909 | 11 |
| PRL | 2.585062 | 0.001092 | 0.386838 | 0.472727 | 11 |
| IL2RB | 2.742739 | 0.000934 | 0.364599 | 0.644444 | 10 |
| F2RL2 | 2.73029 | 0.003432 | 0.366261 | 0.527778 | 9 |
| TWIST2 | 2.809129 | 0.000309 | 0.355982 | 0.527778 | 9 |
| TRIO | 2.622407 | 0.008446 | 0.381329 | 0.535714 | 8 |
| EPS15 | 2.705394 | 0.002056 | 0.369632 | 0.642857 | 8 |
| Name | Description | Average Shortest Path Length | Betweenness Centrality | Closeness Centrality | Neighborhood Connectivity | Node Size | No. of Genes | Adjusted p-Value |
|---|---|---|---|---|---|---|---|---|
| 65007 | biological regulation | 3.72 | 0.138077 | 0.268817 | 8.333333 | 16.12452 | 65 | 0.00348 |
| 50789 | regulation of the biological process | 2.68254 | 0.263324 | 0.372781 | 4.090909 | 15.87451 | 63 | 0.0027 |
| 50794 | regulation of the cellular process | 2.605263 | 0.131545 | 0.383838 | 4.2 | 15.74802 | 62 | 0.0024 |
| 19222 | regulation of the metabolic process | 2.125 | 0.039602 | 0.470588 | 4.75 | 12.49 | 39 | 0.0216 |
| 31323 | regulation of the cellular metabolic process | 0 | 0 | 0 | 7 | 12 | 36 | 0.0449 |
| 23052 | signaling | 2.625 | 0.012372 | 0.380952 | 5.666667 | 12 | 36 | 0.00789 |
| 32502 | developmental process | 2.858824 | 0.112328 | 0.349794 | 7 | 11.6619 | 34 | 0.0216 |
| 7275 | multicellular organismal development | 3.904762 | 0.175373 | 0.256098 | 5 | 11.31371 | 32 | 0.0216 |
| 10468 | regulation of gene expression | 1.333333 | 0.015726 | 0.75 | 3 | 11.13553 | 31 | 0.0299 |
| 48856 | anatomical structure development | 2.426471 | 0.066427 | 0.412121 | 5.428571 | 10.77033 | 29 | 0.0295 |
| 16043 | cellular component organization | 2.625 | 0.017086 | 0.380952 | 5.2 | 10.77033 | 29 | 0.0207 |
| 48731 | system development | 3.531915 | 0.30602 | 0.283133 | 5.125 | 10.58301 | 28 | 0.0194 |
| 23033 | signaling pathway | 1.5 | 0.007868 | 0.666667 | 2.5 | 10.3923 | 27 | 0.0113 |
| 48869 | cellular developmental process | 2.774194 | 0.143077 | 0.360465 | 6.166667 | 9.591663 | 23 | 0.0143 |
| 48523 | negative regulation of cellular process | 3 | 0.074143 | 0.333333 | 5.25 | 9.380832 | 22 | 0.0371 |
| 30154 | cell differentiation | 2.615385 | 0.069689 | 0.382353 | 4 | 9.380832 | 22 | 0.0184 |
| 48513 | organ development | 2.827586 | 0.199668 | 0.353659 | 5.375 | 9.165151 | 21 | 0.0482 |
| 7166 | cell surface receptor linked signaling pathway | 1 | 0.005863 | 1 | 1.5 | 8.944272 | 20 | 0.00875 |
| 51239 | regulation of the multicellular organismal process | 1.9375 | 0.058335 | 0.516129 | 4.285714 | 8.485281 | 18 | 0.0083 |
| 35466 | regulation of signaling pathway | 0 | 0 | 0 | 11 | 7.745967 | 15 | 0.0299 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guha, P.; Roychoudhury, S.; Singha, S.; Kalita, J.C.; Kolesarova, A.; Jamal, Q.M.S.; Jha, N.K.; Kumar, D.; Ruokolainen, J.; Kesari, K.K. A Comparative Cross-Platform Meta-Analysis to Identify Potential Biomarker Genes Common to Endometriosis and Recurrent Pregnancy Loss. Appl. Sci. 2021, 11, 3349. https://doi.org/10.3390/app11083349
Guha P, Roychoudhury S, Singha S, Kalita JC, Kolesarova A, Jamal QMS, Jha NK, Kumar D, Ruokolainen J, Kesari KK. A Comparative Cross-Platform Meta-Analysis to Identify Potential Biomarker Genes Common to Endometriosis and Recurrent Pregnancy Loss. Applied Sciences. 2021; 11(8):3349. https://doi.org/10.3390/app11083349
Chicago/Turabian StyleGuha, Pokhraj, Shubhadeep Roychoudhury, Sobita Singha, Jogen C. Kalita, Adriana Kolesarova, Qazi Mohammad Sajid Jamal, Niraj Kumar Jha, Dhruv Kumar, Janne Ruokolainen, and Kavindra Kumar Kesari. 2021. "A Comparative Cross-Platform Meta-Analysis to Identify Potential Biomarker Genes Common to Endometriosis and Recurrent Pregnancy Loss" Applied Sciences 11, no. 8: 3349. https://doi.org/10.3390/app11083349
APA StyleGuha, P., Roychoudhury, S., Singha, S., Kalita, J. C., Kolesarova, A., Jamal, Q. M. S., Jha, N. K., Kumar, D., Ruokolainen, J., & Kesari, K. K. (2021). A Comparative Cross-Platform Meta-Analysis to Identify Potential Biomarker Genes Common to Endometriosis and Recurrent Pregnancy Loss. Applied Sciences, 11(8), 3349. https://doi.org/10.3390/app11083349












