PPIA and YWHAZ Constitute a Stable Pair of Reference Genes during Electrical Stimulation in Mesenchymal Stem Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Direct Current ES
2.3. RNA Extraction and Quantitation
2.4. Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
2.5. Metabolic Activity
2.6. Primer Amplification Efficiency
2.7. Data Analysis
3. Results
3.1. Primers Are Effective and Selective for Their Respective Genes
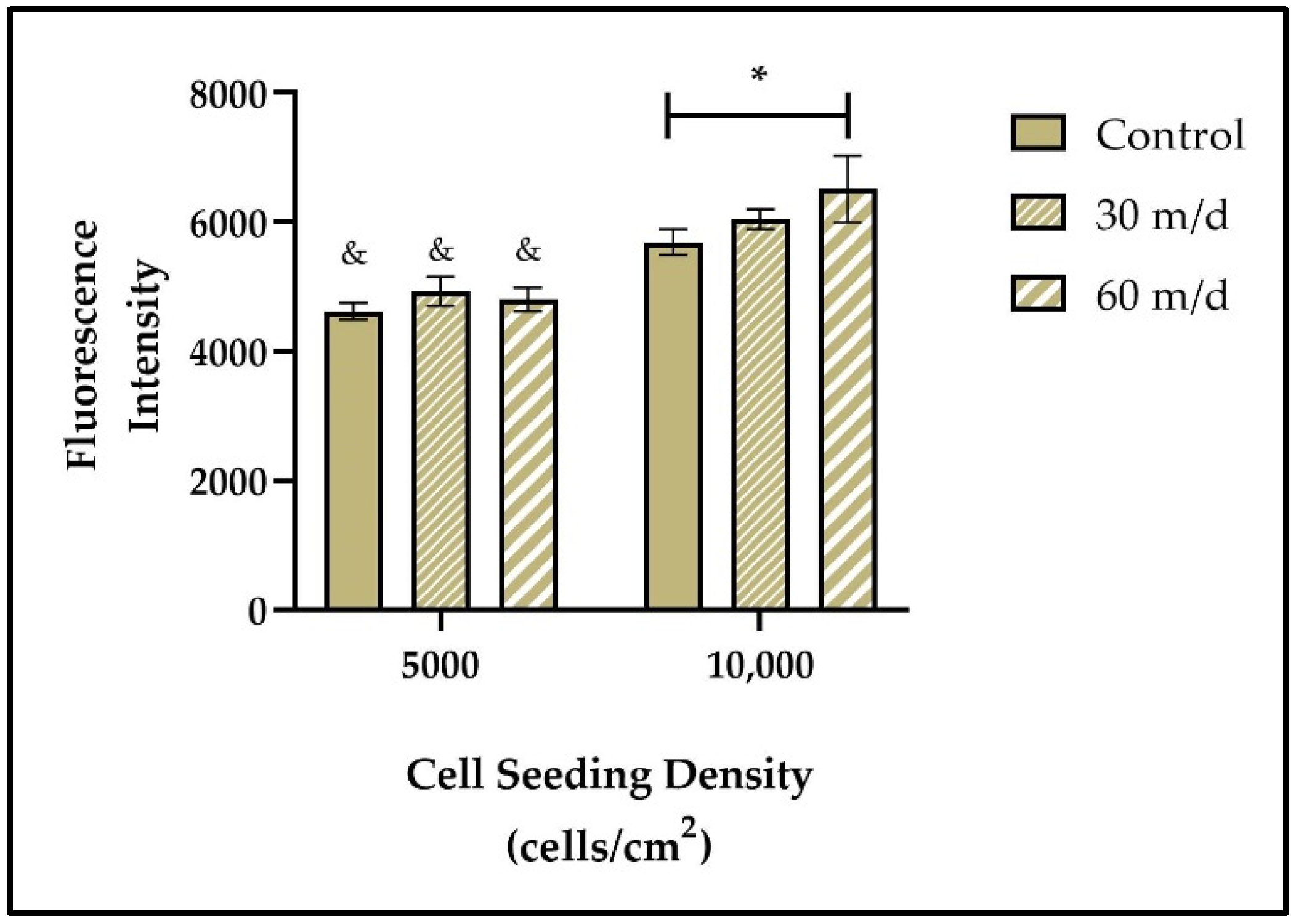
3.2. Cells Remain Viable under ES
3.3. The Effect of the Duration of ES on Reference Gene Activity
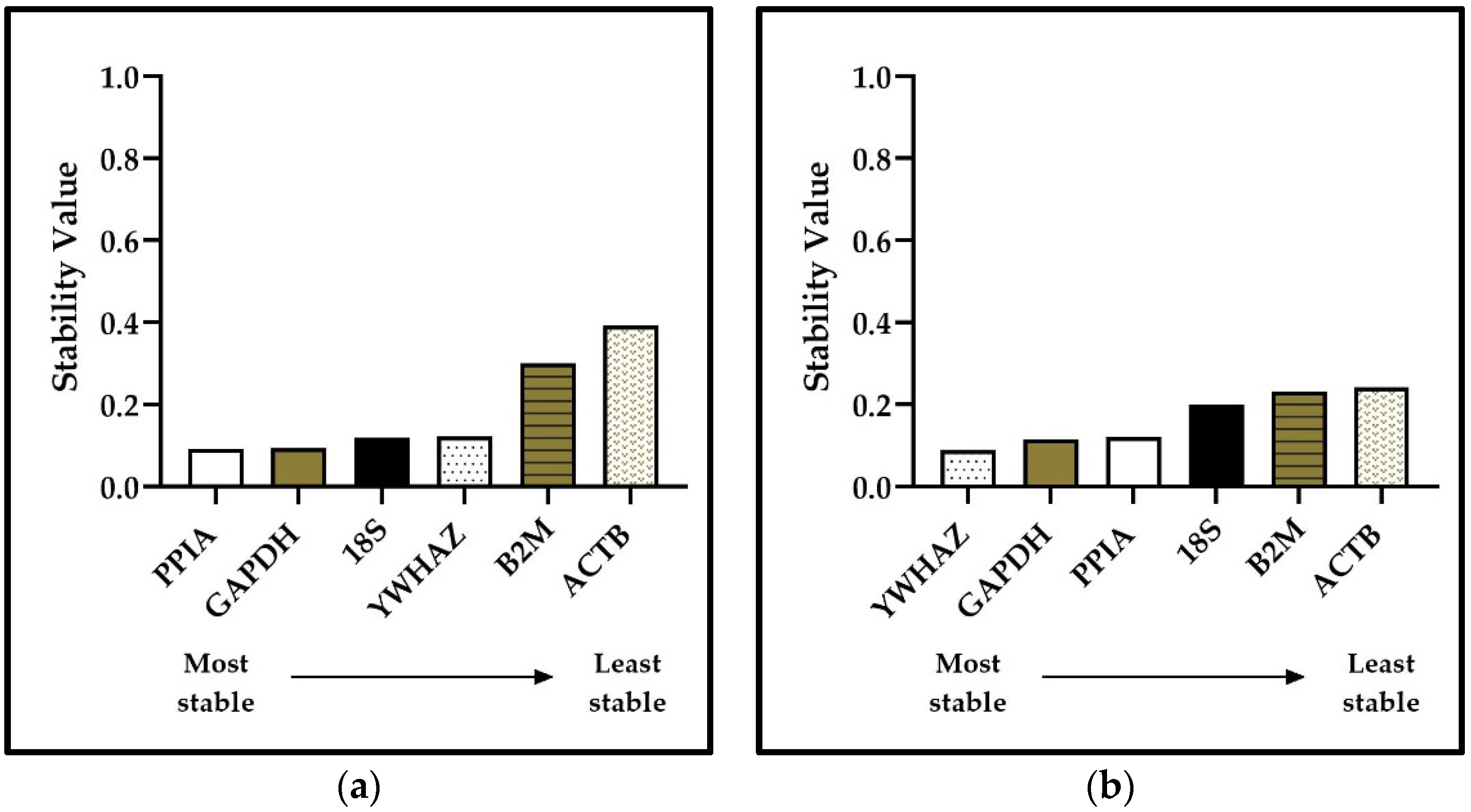
3.4. NormFinder Results
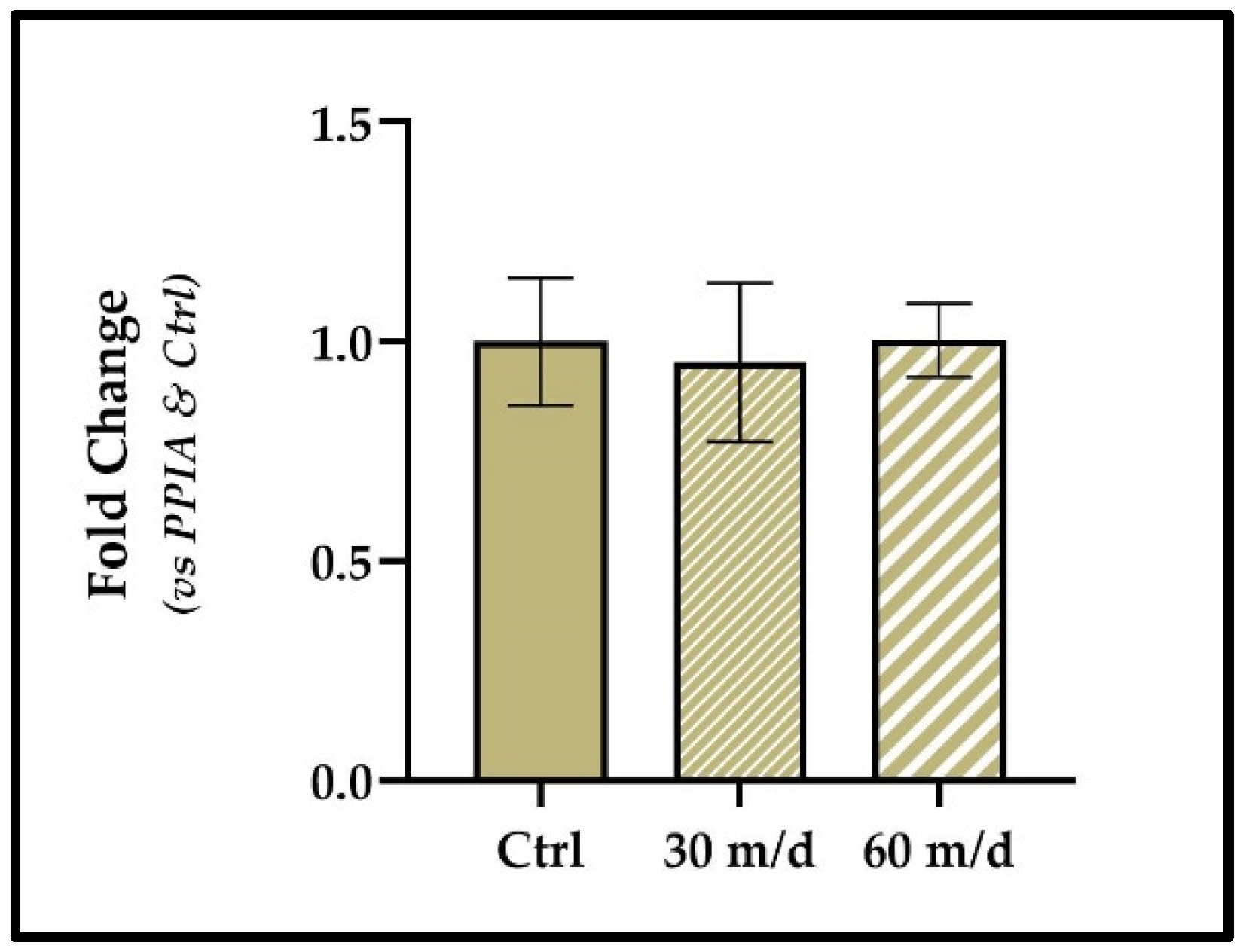
3.5. ∆∆CT Method Results
3.6. BestKeeper Results
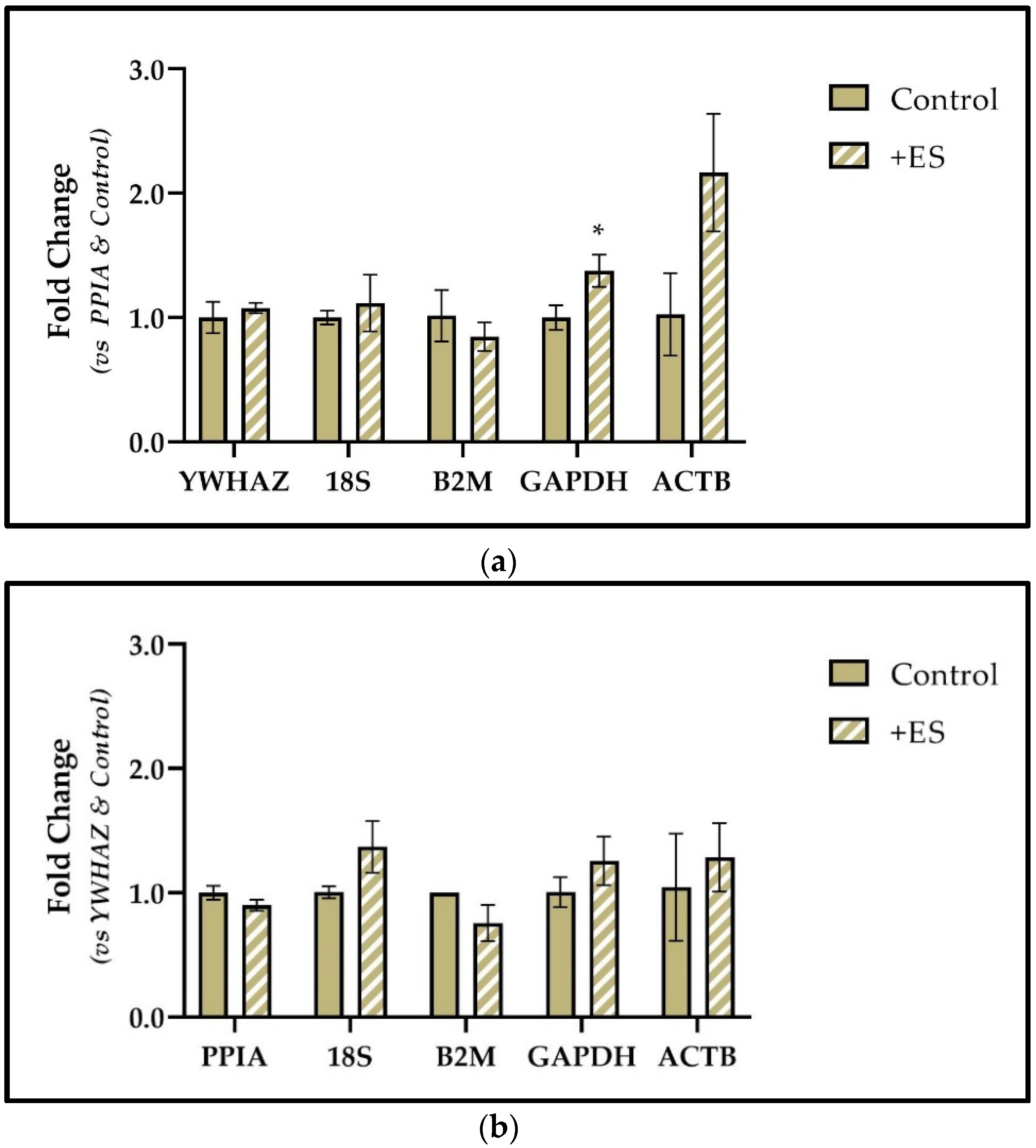
3.7. PPIA and YWHAZ Expression in Primary Human MSCs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Okamura, K.; Inagaki, Y.; Matsui, T.K.; Matsubayashi, M. OPEN RT-qPCR analyses on the osteogenic differentiation from human iPS cells: An investigation of reference genes. Sci. Rep. 2020, 10, 11748. [Google Scholar] [CrossRef]
- Lee, W.J.; Jeon, R.H.; Jang, S.J.; Park, J.S.; Lee, S.C.; Baregundi Subbarao, R.; Lee, S.L.; Park, B.W.; King, W.A.; Rho, G.J. Selection of reference genes for quantitative gene expression in porcine mesenchymal stem cells derived from various sources along with differentiation into multilineages. Stem Cells Int. 2015, 2015, 235192. [Google Scholar] [CrossRef]
- Denu, R.A.; Hematti, P. Effects of Oxidative Stress on Mesenchymal Stem Cell Biology. Oxid. Med. Cell. Longev. 2016, 2016, 2989076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanna, H.; Mir, L.M.; Andre, F.M. In vitro osteoblastic differentiation of mesenchymal stem cells generates cell layers with distinct properties. Stem Cell Res. Ther. 2018, 9, 203. [Google Scholar] [CrossRef]
- Ullah, I.; Subbarao, R.B.; Rho, G.J. Human mesenchymal stem cells-current trends and future prospective. Biosci. Rep. 2015, 35, e00191. [Google Scholar] [CrossRef]
- Li, P.; Xu, J.; Liu, L.; Zhang, Y.; Liu, M.; Fan, Y. Promoting Proliferation and Differentiation of Pre-Osteoblasts MC3T3-E1 Cells Under Combined Mechanical and Electrical Stimulation. J. Biomed. Nanotechnol. 2019, 15, 921–929. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Shi, R.; Liu, Z.; Ouyang, H.; Yu, M.; Zhao, C.; Zou, Y.; Jiang, D.; Zhang, J.; Li, Z. Self-powered implantable electrical stimulator for osteoblasts’ proliferation and differentiation. Nano Energy 2019, 59, 705–714. [Google Scholar] [CrossRef]
- Gu, X.; Fu, J.; Bai, J.; Wang, J.; Pan, W.; Zhang, C. Low-Frequency Electrical Stimulation Induces the Proliferation and Differentiation of Peripheral Blood Stem Cells Into Schwann Cells. Am. J. Med. Sci. 2015, 349, 157–161. [Google Scholar] [CrossRef]
- Lee, J.H.; Jeon, W.-Y.; Kim, H.-H.; Lee, E.-J.; Kim, H.-W. Electrical stimulation by enzymatic biofuel cell to promote proliferation, migration and differentiation of muscle precursor cells. Biomaterials 2015, 53, 358–369. [Google Scholar] [CrossRef]
- Tai, G.; Tai, M.; Zhao, M. Electrically stimulated cell migration and its contribution to wound healing. Burn. Trauma 2018, 6, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, J.F.; Liu, J.; Zhang, L.; Jiang, J.Y.; Russell, M.; Lyeth, B.G.; Nolta, J.A.; Zhao, M. Electrical Guidance of Human Stem Cells in the Rat Brain. Stem Cell Rep. 2017, 9, 177–189. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.Y.; Hou, H.S.; Sun, Y.S.; Cheng, J.Y.; Lo, K.Y. Correlation between cell migration and reactive oxygen species under electric field stimulation. Biomicrofluidics 2015, 9, 054120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Z.; Watt, C.; Karystinou, A.; Roelofs, A.J.; McCaig, C.D.; Gibson, I.R.; De Bari, C. Directed migration of human bone marrow mesenchymal stem cells in a physiological direct current electric field. Eur. Cells Mater. 2011, 22, 344–358. [Google Scholar] [CrossRef]
- Mobini, S.; Leppik, L.; Thottakkattumana Parameswaran, V.; Barker, J.H. In vitro effect of direct current electrical stimulation on rat mesenchymal stem cells. PeerJ 2017, 5, e2821. [Google Scholar] [CrossRef] [Green Version]
- Eischen-Loges, M.; Oliveira, K.M.C.; Bhavsar, M.B.; Barker, J.H.; Leppik, L. Pretreating mesenchymal stem cells with electrical stimulation causes sustained long-lasting pro-osteogenic effects. PeerJ 2018, 6, e4959. [Google Scholar] [CrossRef]
- Srirussamee, K.; Mobini, S.; Cassidy, N.J.; Cartmell, S.H. Direct electrical stimulation enhances osteogenesis by inducing Bmp2 and Spp1 expressions from macrophages and preosteoblasts. Biotechnol. Bioeng. 2019, 116, 3421–3432. [Google Scholar] [CrossRef]
- Balint, R.; Cassidy, N.J.; Hidalgo-Bastida, L.A.; Cartmell, S. Electrical stimulation enhanced mesenchymal stem cell gene expression for orthopaedic tissue repair. J. Biomater. Tissue Eng. 2013, 3, 212–221. [Google Scholar] [CrossRef]
- Nazari, F.; Parham, A.; Maleki, A.F. GAPDH, β-actin and β2-microglobulin, as three common reference genes, are not reliable for gene expression studies in equine adipose- and marrow-derived mesenchymal stem cells. J. Anim. Sci. Technol. 2015, 57, 18. [Google Scholar] [CrossRef] [Green Version]
- Jeon, R.-H.; Lee, W.-J.; Son, Y.-B.; Bharti, D.; Shivakumar, S.B.; Lee, S.-L.; Rho, G.-J. PPIA, HPRT1, and YWHAZ Genes Are Suitable for Normalization of mRNA expression in Long-Term Expanded Human Mesenchymal Stem Cells. Biomed Res. Int. 2019, 2019, 3093545. [Google Scholar] [CrossRef]
- Ayanoğlu, F.B.; Elçin, A.E.; Elçin, Y.M. Evaluation of the stability of standard reference genes of adipose-derived mesenchymal stem cells during in vitro proliferation and differentiation. Mol. Biol. Rep. 2020, 47, 2109–2122. [Google Scholar] [CrossRef] [PubMed]
- Quiroz, F.G.; Posada, O.M.; Gallego-Perez, D.; Higuita-Castro, N.; Sarassa, C.; Hansford, D.J.; Agudelo-Florez, P.; López, L.E. Housekeeping gene stability influences the quantification of osteogenic markers during stem cell differentiation to the osteogenic lineage. Cytotechnology 2010, 62, 109–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vigano, M.; Orfei, C.P.; de Girolamo, L.; Pearson, J.R.; Ragni, E.; De Luca, P.; Colombini, A. Housekeeping Gene Stability in Human Mesenchymal Stem and Tendon Cells Exposed to Tenogenic Factors. Tissue Eng. Part C 2018, 24, 360–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- James, S.; Fox, J.; Afsari, F.; Lee, J.; Clough, S.; Knight, C.; Ashmore, J.; Ashton, P.; Preham, O.; Hoogduijn, M.; et al. Multiparameter Analysis of Human Bone Marrow Stromal Cells Identifies Distinct Immunomodulatory and Differentiation-Competent Subtypes. Stem Cell Rep. 2015, 4, 1004–1015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [Green Version]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Li, X.; Yang, Q.; Bai, J.; Xuan, Y.; Wang, Y. Identification of appropriate reference genes for human mesenchymal stem cell analysis by quantitative real-time PCR. Biotechnol. Lett. 2015, 37, 67–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwaber, J.; Andersen, S.; Nielsen, L. Shedding light: The importance of reverse transcription efficiency standards in data interpretation. Biomol. Detect. Quantif. 2019, 17, 100077. [Google Scholar] [CrossRef] [PubMed]
- Sundaram, V.; Sampathkumar, N.; Massaad, C.; Grenier, J. Optimal use of statistical methods to validate reference gene stability in longitudinal studies. PLoS ONE 2019, 14, e0219440. [Google Scholar] [CrossRef] [Green Version]
- Jacob, F.; Guertler, R.; Naim, S.; Nixdorf, S.; Fedier, A.; Hacker, N.F.; Heinzelmann-Schwarz, V. Careful Selection of Reference Genes Is Required for Reliable Performance of RT-qPCR in Human Normal and Cancer Cell Lines. PLoS ONE 2013, 8, e59180. [Google Scholar] [CrossRef] [Green Version]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Svec, D.; Tichopad, A.; Novosadova, V.; Pfaffl, M.W.; Kubista, M. How good is a PCR efficiency estimate: Recommendations for precise and robust qPCR efficiency assessments. Biomol. Detect. Quantif. 2015, 3, 9–16. [Google Scholar] [CrossRef] [Green Version]
- Thrivikraman, G.; Madras, G.; Basu, B. Electrically driven intracellular and extracellular nanomanipulators evoke neurogenic/cardiomyogenic differentiation in human mesenchymal stem cells. Biomaterials 2016, 77, 26–43. [Google Scholar] [CrossRef]
- Clark, C.C.; Wang, W.; Brighton, C.T. Up-regulation of expression of selected genes in human bone cells with specific capacitively coupled electric fields. J. Orthop. Res. 2014, 32, 894–903. [Google Scholar] [CrossRef]
- Ravikumar, K.; Boda, S.K.; Basu, B. Synergy of substrate conductivity and intermittent electrical stimulation towards osteogenic differentiation of human mesenchymal stem cells. Bioelectrochemistry 2017, 116, 52–64. [Google Scholar] [CrossRef]
- McCaig, C.D.; Rajnicek, A.M.; Song, B.; Zhao, M. Controlling Cell Behavior Electrically: Current Views and Future Potential. Physiol. Rev. 2005, 85, 943–978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruzgys, P.; Novickij, V.; Novickij, J.; Šatkauskas, S. Influence of the electrode material on ROS generation and electroporation efficiency in low and high frequency nanosecond pulse range. Bioelectrochemistry 2019, 127, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Tristan, C.; Shahani, N.; Sedlak, T.W.; Sawa, A. The diverse functions of GAPDH: Views from different subcellular compartments. Cell. Signal. 2011, 23, 317–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bunnell, T.M.; Burbach, B.J.; Shimizu, Y.; Ervasti, J.M. β-Actin specifically controls cell growth, migration, and the G-actin pool. Mol. Biol. Cell 2011, 22, 4047–4058. [Google Scholar] [CrossRef]
- Kim, I.S.; Song, J.K.; Song, Y.M.; Cho, T.H.; Lee, T.H.; Lim, S.S.; Kim, S.J.; Hwang, S.J. Novel effect of biphasic electric current on in vitro osteogenesis and cytokine production in human mesenchymal stromal cells. Tissue Eng. Part A 2009, 15, 2411–2422. [Google Scholar] [CrossRef] [Green Version]
- Hu, W.W.; Hsu, Y.T.; Cheng, Y.C.; Li, C.; Ruaan, R.C.; Chien, C.C.; Chung, C.A.; Tsao, C.W. Electrical stimulation to promote osteogenesis using conductive polypyrrole films. Mater. Sci. Eng. C 2014, 37, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.J.; Lee, G.S.; Chun, H. Electrical stimulation drives chondrogenesis of mesenchymal stem cells in the absence of exogenous growth factors OPEN. Nat. Publ. Gr. 2016, 6, 39302. [Google Scholar] [CrossRef]
- Qi, Z.; Xia, P.; Pan, S.; Zheng, S.; Fu, C.; Chang, Y.; Ma, Y.; Wang, J.; Yang, X. Combined treatment with electrical stimulation and insulin-like growth factor-1 promotes bone regeneration in vitro. PLoS ONE 2018, 13, e0197006. [Google Scholar] [CrossRef] [Green Version]
- Sun, S.; Liu, Y.; Lipsky, S.; Cho†, M. Physical manipulation of calcium oscillations facilitates osteodifferentiation of human mesenchymal stem cells. FASEB J. 2007, 21, 1472–1480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jing, W.; Zhang, Y.; Cai, Q.; Chen, G.; Wang, L.; Yang, X.; Zhong, W. Study of Electrical Stimulation with Different Electric-Field Intensities in the Regulation of the Differentiation of PC12 Cells. ACS Chem. Neurosci. 2019, 10, 348–357. [Google Scholar] [CrossRef]
- Cai, S.; Bodle, J.C.; Mathieu, P.S.; Amos, A.; Hamouda, M.; Bernacki, S.; Mccarty, G.; Loboa, E.G. Primary cilia are sensors of electrical field stimulation to induce osteogenesis of human adipose-derived stem cells. FASEB J. 2017, 31, 346–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thrivikraman, G.; Madras, G.; Basu, B. Intermittent electrical stimuli for guidance of human mesenchymal stem cell lineage commitment towards neural-like cells on electroconductive substrates. Biomaterials 2014, 35, 6219–6235. [Google Scholar] [CrossRef]
- Nyoung Heo, D.; Acquah, N.; Kim, J.; Lee, S.-J.; Castro, N.J.; Zhang, L.G. Directly Induced Neural Differentiation of Human Adipose-Derived Stem Cells Using Three-Dimensional Culture System of Conductive Microwell with Electrical Stimulation. Tissue Eng. Part A 2018, 24, 537–545. [Google Scholar] [CrossRef]
- Zhang, J.; Li, M.; Kang, E.-T.; Neoh, K.G. Electrical stimulation of adipose-derived mesenchymal stem cells in conductive scaffolds and the roles of voltage-gated ion channels. Acta Biomater. 2016, 32, 46–56. [Google Scholar] [CrossRef]
- Piruzyan, M.; Shitanda, I.; Shimauchi, Y.; Okita, G.; Tsurekawa, Y.; Moriuchi, M.; Nakano, Y.; Teramoto, K.; Suico, M.A.; Shuto, T.; et al. A novel condition of mild electrical stimulation exerts immunosuppression via hydrogen peroxide production that controls multiple signaling pathway. PLoS ONE 2020, 15, e0234867. [Google Scholar] [CrossRef]
- Lee, G.S.; Kim, M.G.; Kwon, H.J. Electrical stimulation induces direct reprogramming of human dermal fibroblasts into hyaline chondrogenic cells. Biochem. Biophys. Res. Commun. 2019, 513, 990–996. [Google Scholar] [CrossRef]
- He, X.; Li, L.; Tang, M.; Zeng, Y.; Li, H.; Yu, X. Biomimetic electrical stimulation induces rat bone marrow mesenchymal stem cells to differentiate into cardiomyocyte-like cells via TGF-beta 1 in vitro. Prog. Biophys. Mol. Biol. 2019, 148, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Zhang, X.; Yu, X.; Wang, S.; Qiu, J.; Tang, W.; Li, L.; Liu, H.; Wang, Z.L. Self-Powered Electrical Stimulation for Enhancing Neural Differentiation of Mesenchymal Stem Cells on Graphene-Poly(3,4-ethylenedioxythiophene) Hybrid Microfibers. ACS Nano 2016, 10, 5086–5095. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Rouabhia, M.; Zhang, Z. Pulsed electrical stimulation benefits wound healing by activating skin fibroblasts through the TGFβ1/ERK/NF-κB axis. Biochim. Biophys. Acta Gen. Subj. 2016, 1860, 1551–1559. [Google Scholar] [CrossRef]
- Zhuang, H.; Wang, W.; Seldes, R.M.; David Tahernia, A.; Fan, H.; Brighton, C.T. Electrical Stimulation Induces the Level of TGF-beta1 mRNA in Osteoblastic Cells by a Mechanism Involving Calcium/Calmodulin Pathway. Biochem. Biophys. Res. Commun. 1997, 237, 225–229. [Google Scholar] [CrossRef]
- Hiemer, B.; Krogull, M.; Bender, T.; Ziebart, J.; Krueger, S.; Bader, R.; Jonitz-Heincke, A. Effect of electric stimulation on human chondrocytes and mesenchymal stem cells under normoxia and hypoxia. Mol. Med. Rep. 2018, 18, 2133–2141. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Gene Name | Forward Sequence | Reverse Sequence | No. of Transcript Variants Detected, (Total) |
|---|---|---|---|
| Peptidylprolyl isomerase A (PPIA) | TGCTGGACCCAACACAAATG | AACACCACATGCTTGCCATC | 2, (2) |
| Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta (YWHAZ) | CGAAGCTGAAGCAGGAGAAG | TTTGTGGGACAGCATGGATG | 6, (6) |
| 18S ribosomal RNA (18S) | CTCAACACGGGAAACCTCAC | CGCTCCACCAACTAAGAACG | 1, (1) |
| Beta-2-microglobulin (B2M) | TGGGTTTCATCCATCCGACA | GCTTACATGTCTCGATCCCACT | 1, (1) |
| Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | AAGGTCGGAGTCAACGGATT | CTCCTGGAAGATGGTGATGG | 5, (6) |
| Beta-Actin (ACTB) | CACAGAGCCTCGCCTTTGC | CCATCACGCCCTGGTGC | 1, (1) |
| Gene Name | Reference | Product Length | Slope | Efficiency (%) | y-Intercept | R2 Value |
|---|---|---|---|---|---|---|
| PPIA | NM_001300981.2 | 81 | −3.184 | 106.10 | 27.07 | 0.999 |
| YWHAZ | NM_001135699.2 | 110 | −3.250 | 103.09 | 26.45 | 0.999 |
| 18S | Universal Probe Library, 18S probe 77 | 114 | −3.184 | 106.10 | 15.67 | 0.999 |
| B2M | NM_004048.4 | 219 | −3.311 | 100.46 | 26.69 | 0.997 |
| GAPDH | NM_001357943.2 | 171 | −3.367 | 98.15 | 26.54 | 0.998 |
| ACTB | NM_001101.5 | 189 | −3.495 | 93.25 | 27.72 | 0.999 |
| Pearson’s Correlation Coefficient (r) | ||||
|---|---|---|---|---|
| Reference Gene Pair | Seeding Density (Cells/cm2) | Average | Standard Deviation | |
| 5000 | 10,000 | |||
| PPIA:YWHAZ | 0.979 | 0.994 | 0.987 | 0.0106 |
| PPIA:B2M | 0.990 | 0.942 | 0.966 | 0.0339 |
| YWHAZ:18S | 0.981 | 0.912 | 0.947 | 0.0488 |
| PPIA:GAPDH | 0.932 | 0.957 | 0.945 | 0.0177 |
| YWHAZ:B2M | 0.971 | 0.914 | 0.943 | 0.0403 |
| PPIA:18S | 0.957 | 0.875 | 0.916 | 0.0580 |
| YWHAZ:GAPDH | 0.842 | 0.961 | 0.902 | 0.0841 |
| B2M:GAPDH | 0.923 | 0.822 | 0.873 | 0.0714 |
| 18S:GAPDH | 0.801 | 0.956 | 0.879 | 0.1096 |
| 18S:B2M | 0.930 | 0.68 | 0.805 | 0.1768 |
| GAPDH:ACTB | 0.231 | 0.958 | 0.595 | 0.5141 |
| PPIA:ACTB | −0.111 | 0.883 | 0.386 | 0.7029 |
| B2M:ACTB | −0.110 | 0.723 | 0.307 | 0.5890 |
| YWHAZ:ACTB | −0.271 | 0.869 | 0.299 | 0.8061 |
| 18S:ACTB | −0.363 | 0.880 | 0.259 | 0.8789 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Steel, L.; Ansell, D.M.; Amaya, E.; Cartmell, S.H. PPIA and YWHAZ Constitute a Stable Pair of Reference Genes during Electrical Stimulation in Mesenchymal Stem Cells. Appl. Sci. 2022, 12, 153. https://doi.org/10.3390/app12010153
Steel L, Ansell DM, Amaya E, Cartmell SH. PPIA and YWHAZ Constitute a Stable Pair of Reference Genes during Electrical Stimulation in Mesenchymal Stem Cells. Applied Sciences. 2022; 12(1):153. https://doi.org/10.3390/app12010153
Chicago/Turabian StyleSteel, Lynsey, David M. Ansell, Enrique Amaya, and Sarah H. Cartmell. 2022. "PPIA and YWHAZ Constitute a Stable Pair of Reference Genes during Electrical Stimulation in Mesenchymal Stem Cells" Applied Sciences 12, no. 1: 153. https://doi.org/10.3390/app12010153






