End-to-End Deep Learning CT Image Reconstruction for Metal Artifact Reduction
Abstract
:1. Introduction
2. Materials and Methods
2.1. iCTU-Net
2.2. Reference MAR Networks
2.3. Data Generation
2.4. Training
2.5. Evaluation
2.6. Experiments
3. Results
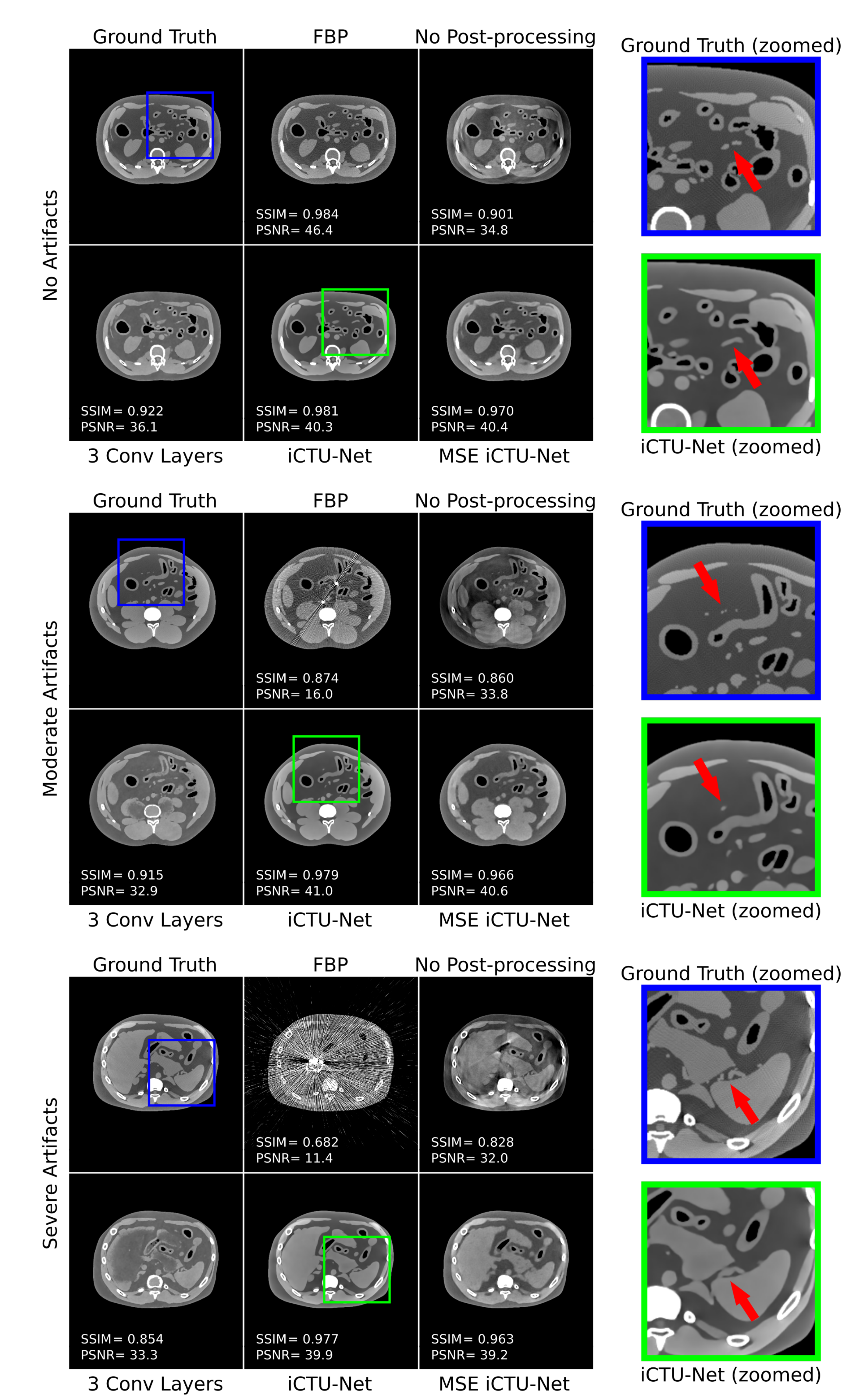
3.1. Ablation Study
3.2. Input Study
3.3. Comparison Study
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Boas, F.E.; Fleischmann, D. CT artifacts: Causes and reduction techniques. Imaging Med. 2012, 4, 229–240. [Google Scholar] [CrossRef] [Green Version]
- Do, T.D.; Heim, J.; Skornitzke, S.; Melzig, C.; Vollherbst, D.F.; Faerber, M.; Pereira, P.L.; Kauczor, H.U.; Sommer, C.M. Single-energy versus dual-energy imaging during CT-guided biopsy using dedicated metal artifact reduction algorithm in an in vivo pig model. PLoS ONE 2021, 16, e0249921. [Google Scholar] [CrossRef]
- McWilliams, S.R.; Murphy, K.P.; Golestaneh, S.; O’Regan, K.N.; Arellano, R.S.; Maher, M.M.; O’Connor, O.J. Reduction of guide needle streak artifact in CT-guided biopsy. J. Vasc. Interv. Radiol. 2014, 25, 1929–1935. [Google Scholar] [CrossRef]
- Stattaus, J.; Kuehl, H.; Ladd, S.; Schroeder, T.; Antoch, G.; Baba, H.A.; Barkhausen, J.; Forsting, M. CT-guided biopsy of small liver lesions: Visibility, artifacts, and corresponding diagnostic accuracy. Cardiovasc. Interv. Radiol. 2007, 30, 928–935. [Google Scholar] [CrossRef]
- Laukamp, K.R.; Zopfs, D.; Wagner, A.; Lennartz, S.; Pennig, L.; Borggrefe, J.; Ramaiya, N.; Hokamp, N.G. CT artifacts from port systems: Virtual monoenergetic reconstructions from spectral-detector CT reduce artifacts and improve depiction of surrounding tissue. Eur. J. Radiol. 2019, 121, 108733. [Google Scholar] [CrossRef]
- Kim, C.; Kim, D.; Lee, K.Y.; Kim, H.; Cha, J.; Choo, J.Y.; Cho, P.K. The optimal energy level of virtual monochromatic images from spectral CT for reducing beam-hardening artifacts due to contrast media in the thorax. Am. J. Roentgenol. 2018, 211, 557–563. [Google Scholar] [CrossRef]
- Lin, M.; Loffroy, R.; Noordhoek, N.; Taguchi, K.; Radaelli, A.; Blijd, J.; Balguid, A.; Geschwind, J.F. Evaluating tumors in transcatheter arterial chemoembolization (TACE) using dual-phase cone-beam CT. Minim. Invasive Ther. Allied Technol. 2011, 20, 276–281. [Google Scholar] [CrossRef]
- Jeong, S.; Kim, S.H.; Hwang, E.J.; Shin, C.i.; Han, J.K.; Choi, B.I. Usefulness of a metal artifact reduction algorithm for orthopedic implants in abdominal CT: Phantom and clinical study results. Am. J. Roentgenol. 2015, 204, 307–317. [Google Scholar] [CrossRef]
- Bismark, R.N.; Frysch, R.; Abdurahman, S.; Beuing, O.; Blessing, M.; Rose, G. Reduction of beam hardening artifacts on real C-arm CT data using polychromatic statistical image reconstruction. Z. Med. Phys. 2020, 30, 40–50. [Google Scholar] [CrossRef]
- Bamberg, F.; Dierks, A.; Nikolaou, K.; Reiser, M.F.; Becker, C.R.; Johnson, T.R. Metal artifact reduction by dual energy computed tomography using monoenergetic extrapolation. Eur. Radiol. 2011, 21, 1424–1429. [Google Scholar] [CrossRef]
- Kalender, W.A.; Hebel, R.; Ebersberger, J. Reduction of CT artifacts caused by metallic implants. Radiology 1987, 164, 576–577. [Google Scholar] [CrossRef]
- Meyer, E.; Raupach, R.; Lell, M.; Schmidt, B.; Kachelrieß, M. Normalized metal artifact reduction (NMAR) in computed tomography. Med. Phys. 2010, 37, 5482–5493. [Google Scholar] [CrossRef]
- Lundervold, A.S.; Lundervold, A. An overview of deep learning in medical imaging focusing on MRI. Z. Med. Phys. 2019, 29, 102–127. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, H. Convolutional neural network based metal artifact reduction in X-ray computed tomography. IEEE Trans. Med Imaging 2018, 37, 1370–1381. [Google Scholar] [CrossRef]
- Ghani, M.U.; Karl, W.C. Deep learning based sinogram correction for metal artifact reduction. Electron. Imaging 2018, 2018, 4721–4728. [Google Scholar] [CrossRef]
- Ghani, M.U.; Karl, W.C. Fast enhanced CT metal artifact reduction using data domain deep learning. IEEE Trans. Comput. Imaging 2019, 6, 181–193. [Google Scholar] [CrossRef] [Green Version]
- Gjesteby, L.; Yang, Q.; Xi, Y.; Zhou, Y.; Zhang, J.; Wang, G. Deep learning methods to guide CT image reconstruction and reduce metal artifacts. In Medical Imaging 2017: Physics of Medical Imaging; International Society for Optics and Photonics: Bellingham, WA, USA, 2017; Volume 10132, p. 101322W. [Google Scholar]
- Lossau, T.; Nickisch, H.; Wissel, T.; Morlock, M.; Grass, M. Learning metal artifact reduction in cardiac CT images with moving pacemakers. Med Image Anal. 2020, 61, 101655. [Google Scholar] [CrossRef]
- Gjesteby, L.; Yang, Q.; Xi, Y.; Claus, B.; Jin, Y.; De Man, B.; Wang, G. Reducing metal streak artifacts in CT images via deep learning: Pilot results. In Proceedings of the 14th International Meeting on Fully Three-Dimensional Image Reconstruction in Radiology and Nuclear Medicine, Xi’an, China, 18–23 June 2017; Volume 14, pp. 611–614. [Google Scholar]
- Wang, J.; Zhao, Y.; Noble, J.H.; Dawant, B.M. Conditional generative adversarial networks for metal artifact reduction in CT images of the ear. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Granada, Spain, 16–20 September 2018; pp. 3–11. [Google Scholar]
- Huang, X.; Wang, J.; Tang, F.; Zhong, T.; Zhang, Y. Metal artifact reduction on cervical CT images by deep residual learning. Biomed. Eng. Online 2018, 17, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Liao, H.; Lin, W.A.; Zhou, S.K.; Luo, J. ADN: Artifact disentanglement network for unsupervised metal artifact reduction. IEEE Trans. Med. Imaging 2019, 39, 634–643. [Google Scholar] [CrossRef] [Green Version]
- Nakao, M.; Imanishi, K.; Ueda, N.; Imai, Y.; Kirita, T.; Matsuda, T. Regularized three-dimensional generative adversarial nets for unsupervised metal artifact reduction in head and neck CT images. IEEE Access 2020, 8, 109453–109465. [Google Scholar] [CrossRef]
- Lee, J.; Gu, J.; Ye, J.C. Unsupervised CT Metal Artifact Learning using Attention-guided β-CycleGAN. IEEE Trans. Med Imaging 2021, 40, 3932–3944. [Google Scholar] [CrossRef]
- Lin, W.A.; Liao, H.; Peng, C.; Sun, X.; Zhang, J.; Luo, J.; Chellappa, R.; Zhou, S.K. Dudonet: Dual domain network for ct metal artifact reduction. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–19 June 2019; pp. 10512–10521. [Google Scholar]
- Li, Y.; Li, K.; Zhang, C.; Montoya, J.; Chen, G.H. Learning to reconstruct computed tomography images directly from sinogram data under a variety of data acquisition conditions. IEEE Trans. Med Imaging 2019, 38, 2469–2481. [Google Scholar] [CrossRef]
- Maas, A.L.; Hannun, A.Y.; Ng, A.Y. Rectifier nonlinearities improve neural network acoustic models. Proc. icml 2013, 30, 3. [Google Scholar]
- Leuschner, J.; Schmidt, M.; Ganguly, P.S.; Andriiashen, V.; Coban, S.B.; Denker, A.; Bauer, D.; Hadjifaradji, A.; Batenburg, K.J.; Maass, P.; et al. Quantitative Comparison of Deep Learning-Based Image Reconstruction Methods for Low-Dose and Sparse-Angle CT Applications. J. Imaging 2021, 7, 44. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Segars, W.P.; Sturgeon, G.; Mendonca, S.; Grimes, J.; Tsui, B.M.W. 4D XCAT phantom for multimodality imaging research. Med. Phys. 2010, 37, 4902–4915. [Google Scholar] [CrossRef]
- Williamson, J.F.; Whiting, B.R.; Benac, J.; Murphy, R.J.; Blaine, G.J.; O’Sullivan, J.A.; Politte, D.G.; Snyder, D.L. Prospects for quantitative computed tomography imaging in the presence of foreign metal bodies using statistical image reconstruction. Med. Phys. 2002, 29, 2404–2418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poludniowski, G.; Landry, G.; Deblois, F.; Evans, P.; Verhaegen, F. SpekCalc: A program to calculate photon spectra from tungsten anode X-ray tubes. Phys. Med. Biol. 2009, 54, N433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leuschner, J.; Schmidt, M.; Baguer, D.O.; Maaß, P. The lodopab-ct dataset: A benchmark dataset for low-dose ct reconstruction methods. arXiv 2019, arXiv:1910.01113. [Google Scholar]
- La Rivière, P.J.; Bian, J.; Vargas, P.A. Penalized-likelihood sinogram restoration for computed tomography. IEEE Trans. Med Imaging 2006, 25, 1022–1036. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Bauer, D.F.; Russ, T.; Waldkirch, B.I.; Tönnes, C.; Segars, W.P.; Schad, L.R.; Zöllner, F.G.; Golla, A.K. Generation of annotated multimodal ground truth datasets for abdominal medical image registration. Int. J. Comput. Assist. Radiol. Surg. 2021, 16, 1277–1285. [Google Scholar] [CrossRef] [PubMed]
- Arabi, H.; Zaidi, H. Deep learning–based metal artefact reduction in PET/CT imaging. Eur. Radiol. 2021, 31, 6384–6396. [Google Scholar] [CrossRef] [PubMed]
- Nasirudin, R.A.; Mei, K.; Panchev, P.; Fehringer, A.; Pfeiffer, F.; Rummeny, E.J.; Fiebich, M.; Noël, P.B. Reduction of metal artifact in single photon-counting computed tomography by spectral-driven iterative reconstruction technique. PLoS ONE 2015, 10, e0124831. [Google Scholar]
- Willemink, M.J.; Persson, M.; Pourmorteza, A.; Pelc, N.J.; Fleischmann, D. Photon-counting CT: Technical principles and clinical prospects. Radiology 2018, 289, 293–312. [Google Scholar] [CrossRef] [PubMed]





| FBP | No Post- | 3 Conv | iCTU-Net | MSE | ||
|---|---|---|---|---|---|---|
| Processing | Layers | iCTU-Net | ||||
| No | SSIM | 0.988±0.020 | ||||
| Artifacts | PSNR | 50.3±4.9 | ||||
| Moderate | SSIM | 0.976±0.007 | ||||
| Artifacts | PSNR | 39.5±1.8 | ||||
| Severe | SSIM | 0.970±0.009 | ||||
| Artifacts | PSNR | 40.7±1.6 | ||||
| All | SSIM | 0.975±0.008 | ||||
| Images | PSNR | 39.5±1.9 |
| FBP | No Metal Input | Metal Input | iCTU-Net | ||
|---|---|---|---|---|---|
| No | SSIM | 0.988±0.020 | |||
| Artifacts | PSNR | 50.3±4.9 | |||
| Moderate | SSIM | 0.976±0.007 | |||
| Artifacts | PSNR | 40.2±1.5 | |||
| Severe | SSIM | 0.970±0.009 | |||
| Artifacts | PSNR | 40.7±0.8 | 40.7±1.6 | ||
| All | SSIM | 0.975±0.008 | |||
| Images | PSNR | 40.0±1.6 |
| FBP | NMAR | U-Net Sino | U-Net Image | iCTU-Net | ||
|---|---|---|---|---|---|---|
| No | SSIM | 0.993±0.010 | ||||
| Artifacts | PSNR | 50.3±4.9 | 50.3±4.9 | |||
| Moderate | SSIM | 0.983±0.011 | ||||
| Artifacts | PSNR | 44.3±3.1 | ||||
| Severe | SSIM | 0.970±0.009 | ||||
| Artifacts | PSNR | 40.7±1.6 | ||||
| All | SSIM | 0.978±0.019 | ||||
| Images | PSNR | 44.1±4.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bauer, D.F.; Ulrich, C.; Russ, T.; Golla, A.-K.; Schad, L.R.; Zöllner, F.G. End-to-End Deep Learning CT Image Reconstruction for Metal Artifact Reduction. Appl. Sci. 2022, 12, 404. https://doi.org/10.3390/app12010404
Bauer DF, Ulrich C, Russ T, Golla A-K, Schad LR, Zöllner FG. End-to-End Deep Learning CT Image Reconstruction for Metal Artifact Reduction. Applied Sciences. 2022; 12(1):404. https://doi.org/10.3390/app12010404
Chicago/Turabian StyleBauer, Dominik F., Constantin Ulrich, Tom Russ, Alena-Kathrin Golla, Lothar R. Schad, and Frank G. Zöllner. 2022. "End-to-End Deep Learning CT Image Reconstruction for Metal Artifact Reduction" Applied Sciences 12, no. 1: 404. https://doi.org/10.3390/app12010404
APA StyleBauer, D. F., Ulrich, C., Russ, T., Golla, A.-K., Schad, L. R., & Zöllner, F. G. (2022). End-to-End Deep Learning CT Image Reconstruction for Metal Artifact Reduction. Applied Sciences, 12(1), 404. https://doi.org/10.3390/app12010404






