Image-Based Automatic Individual Identification of Fish without Obvious Patterns on the Body (Scale Pattern)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Animal
2.1.1. European Seabass
2.1.2. Common Carp
2.2. Experimental Setup and Data Sets
Common Carp
2.3. Data Processing: Identification Procedure
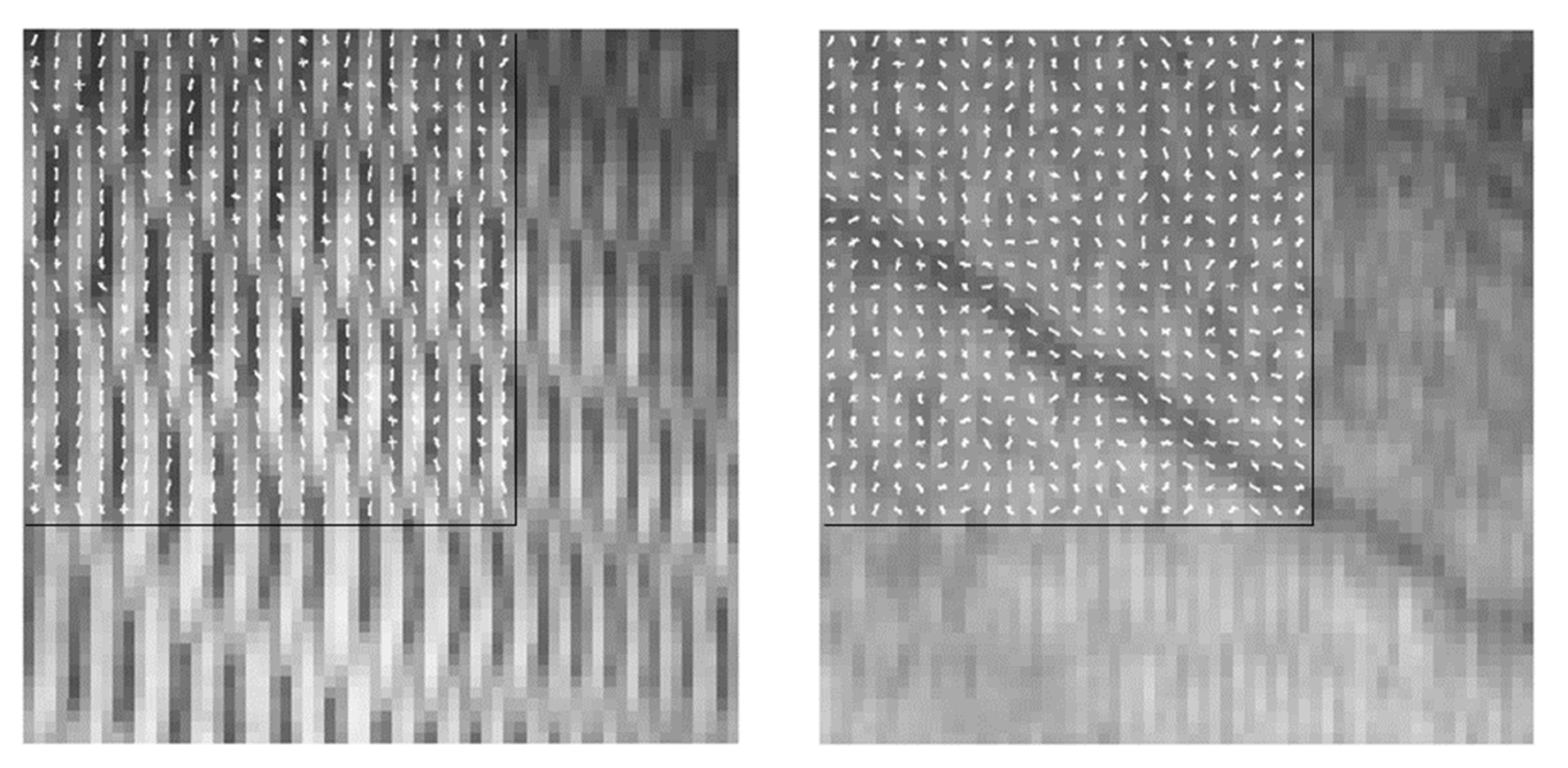
2.3.1. Fish Detection and Feature Extraction
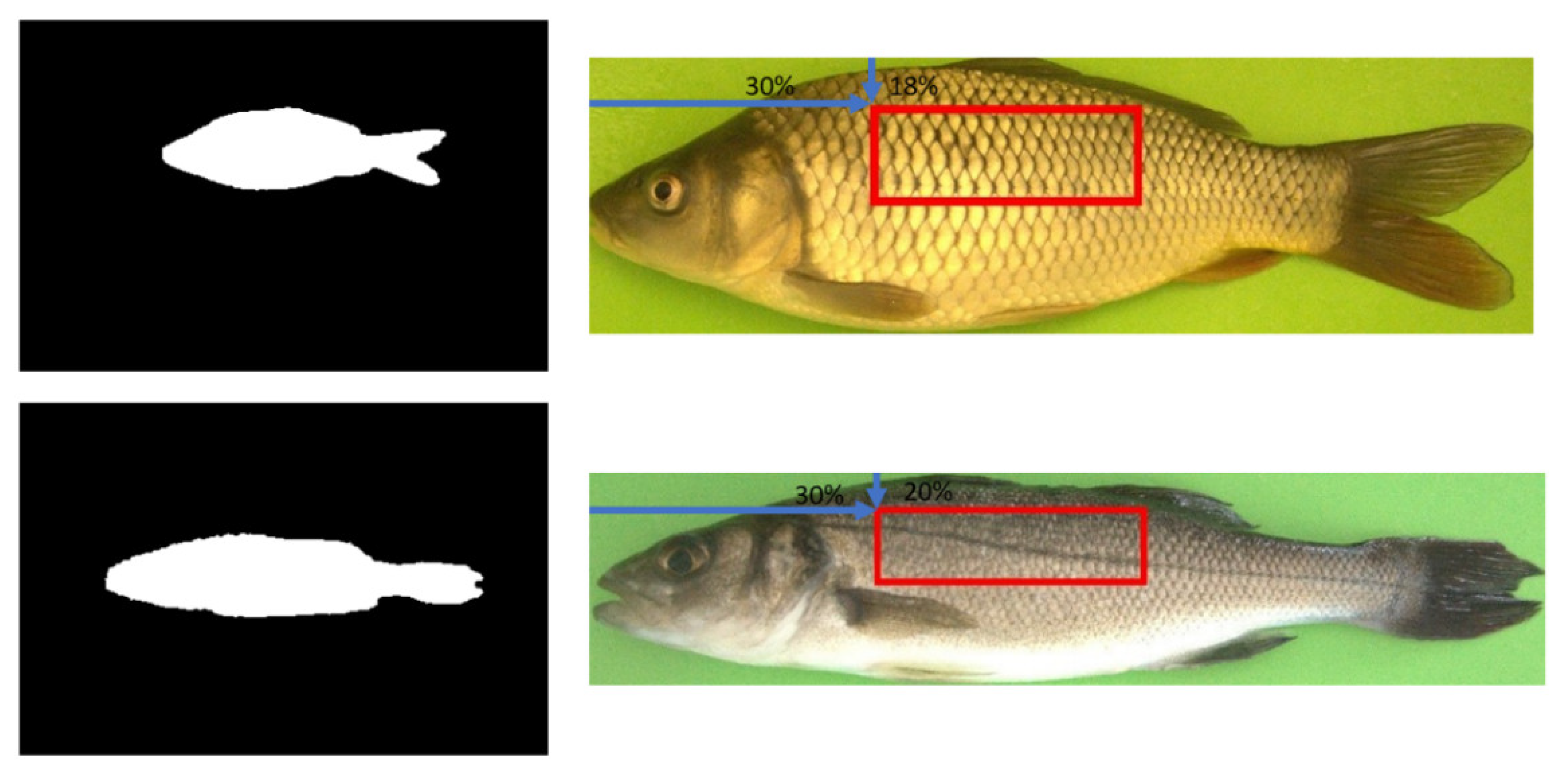
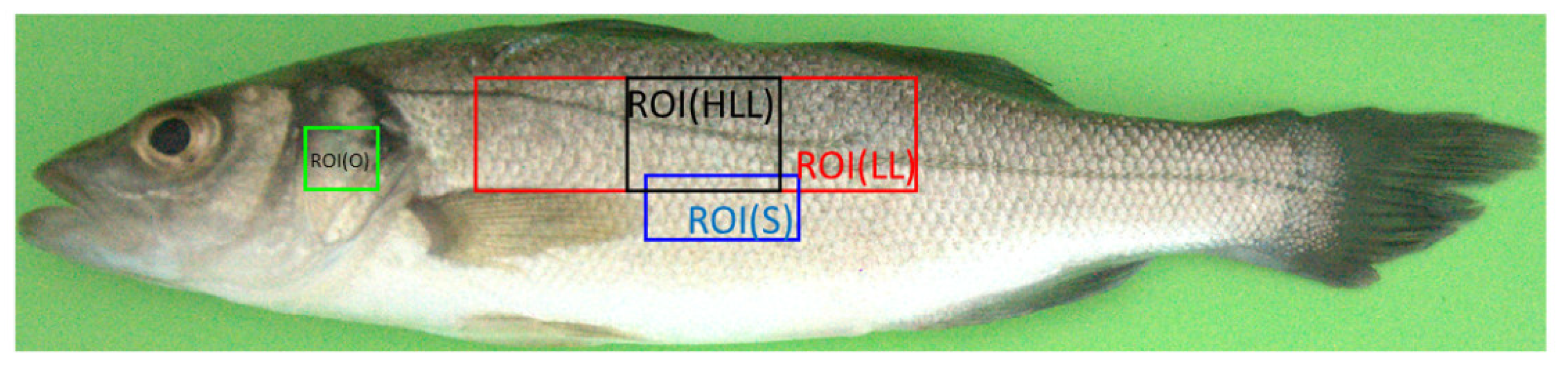
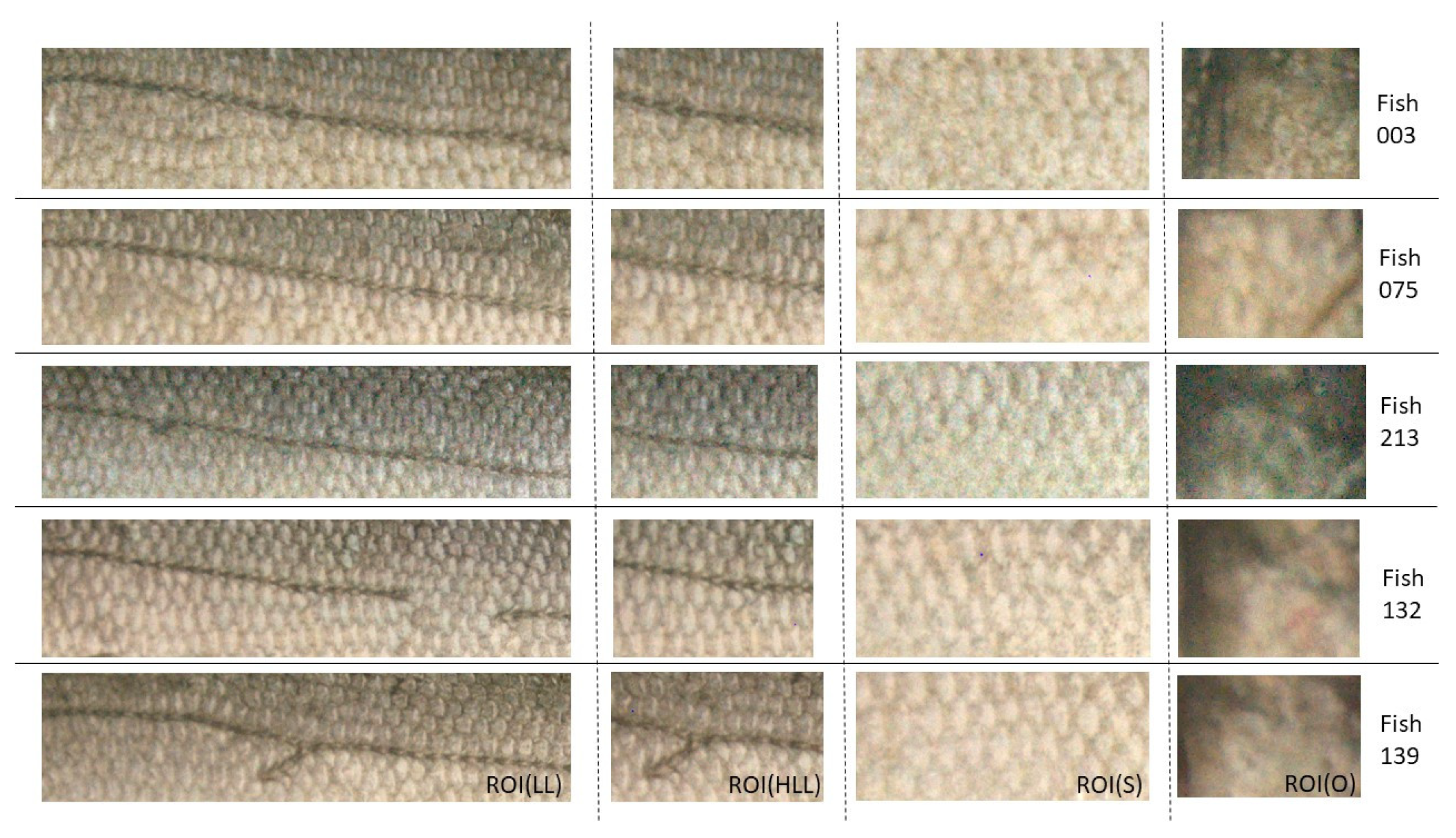
2.3.2. European Seabass ROIs
2.3.3. Common Carp ROIs
2.3.4. Identification Procedure
3. Results
3.1. Seabass Identification Results
3.2. Common Carp Identification Results
4. Discussion
4.1. Seabass
4.2. Carp
5. Conclusions, Limitations, and Future Research
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Føre, M.; Frank, K.; Norton, T.; Svendsen, E.; Alfredsen, J.A.; Dempster, T.; Eguiraun, H.; Watson, W.; Stahl, A.; Sunde, L.M.; et al. Precision fish farming: A new framework to improve production in aquaculture. Biosyst. Eng. 2018, 173, 176–193. [Google Scholar] [CrossRef]
- Bae, M.J.; Park, Y.S. Biological early warning system based on the responses of aquatic organisms to disturbances: A review. Sci. Total Environ. 2014, 466, 635–649. [Google Scholar] [CrossRef] [PubMed]
- Papadakis, V.M.; Papadakis, I.E.; Lamprianidou, F.; Glaropoulos, A.; Kentouri, M. A computer-vision system and methodology for the analysis of fish behavior. Aquac. Eng. 2012, 46, 53–59. [Google Scholar] [CrossRef]
- Khiem, N.M.; Takahashi, Y.; Oanh, D.T.H.; Hai, T.N.; Yasuma, H.; Kimura, N. The use of machine learning to predict acute hepatopancreatic necrosis disease (AHPND) in shrimp farmed on the east coast of the Mekong Delta of Vietnam. Fish. Sci. 2020, 86, 673–683. [Google Scholar] [CrossRef]
- Zhou, C.; Xu, D.; Lin, K.; Sun, C.; Yang, X. Intelligent feeding control methods in aquaculture with an emphasis on fish: A review. Rev. Aquac. 2018, 10, 975–993. [Google Scholar] [CrossRef]
- Li, W.; Ji, Z.; Wang, L.; Sun, C.; Yang, X. Automatic individual identification of Holstein dairy cows using tailhead images. Comput. Electron. Agric. 2017, 142, 622–631. [Google Scholar] [CrossRef]
- Whooley, P.; Berrow, S.; Barnes, C. Photo-identification of fin whales (Balaenoptera physalus L.) off the south coast of Ireland. Mar. Biodivers. Rec. 2011, 4, 1–7. [Google Scholar] [CrossRef]
- Pine, W.E.; Pollock, K.H.; Hightower, J.E.; Kwak, T.J.; Rice, J.A. Management Quantitative Decision Analysis for Sport Fisheries Management. Fisheries 2003, 28, 10–21. [Google Scholar] [CrossRef] [Green Version]
- Cailliet, G.; Mollet, H.; Pittenger, G.; Bedford, D.; Natanson, L. Growth and demography of the Pacific angle shark (Squatina californica), based upon tag returns off California. Mar. Freshw. Res. 1992, 43, 1313. [Google Scholar] [CrossRef]
- Ombredane, D.; Baglinière, J.L.; Marchand, F. The effects of Passive Integrated Transponder tags on survival and growth of juvenile brown trout (Salmo trutta L.) and their use for studying movement in a small river. Hydrobiologia 1998, 371, 99–106. [Google Scholar] [CrossRef]
- Xia, C.; Chon, T.S.; Liu, Y.; Chi, J.; Lee, J.M. Posture tracking of multiple individual fish for behavioral monitoring with visual sensors. Ecol. Inform. 2016, 36, 190–198. [Google Scholar] [CrossRef]
- Casselman, J.M.; Collins, J.J.; Grossman, E.J.; Ihssen, P.E.; Spangler, G.R. Lake Whitefish (Coregonus clupeaformis) stocks of the Ontario waters of Lake Huron. Can. J. Fish. Aquat. Sci. 1981, 38, 1772–1789. [Google Scholar] [CrossRef]
- Cadrin, S.X. Advances in morphometric identification of fishery stocks. Rev. Fish Biol. Fish. 2000, 10, 91–112. [Google Scholar] [CrossRef]
- Saitoh, T.; Shibata, T.; Miyazono, T. Image-based fish recognition. In Proceedings of the 2015 7th International Conference of Soft Computing and Pattern Recognition, SoCPaR 2015, Fukuoka, Japan, 13–15 November 2015; pp. 260–263. [Google Scholar]
- Shafait, F.; Mian, A.; Shortis, M.; Ghanem, B.; Culverhouse, P.F.; Edgington, D.; Cline, D.; Ravanbakhsh, M.; Seager, J.; Harvey, E.S. Fish identification from videos captured in uncontrolled underwater environments. ICES J. Mar. Sci. J. Cons. 2016, 73, 2737–2746. [Google Scholar] [CrossRef]
- Hsiao, Y.H.; Chen, C.C.; Lin, S.I.; Lin, F.P. Real-world underwater fish recognition and identification, using sparse representation. Ecol. Inform. 2014, 23, 13–21. [Google Scholar] [CrossRef]
- Villon, S.; Mouillot, D.; Chaumont, M.; Darling, E.S.; Subsol, G.; Claverie, T.; Villéger, S. A Deep learning method for accurate and fast identification of coral reef fishes in underwater images. Ecol. Inform. 2018, 48, 238–244. [Google Scholar] [CrossRef] [Green Version]
- Villon, S.; Chaumont, M.; Subsol, G.; Villéger, S.; Claverie, T.; Mouillot, D. Coral reef fish detection and recognition in underwater videos by supervised machine learning: Comparison between deep learning and HOG+SVM methods. Lect. Notes Comput. Sci. 2016, 10016, 160–171. [Google Scholar] [CrossRef] [Green Version]
- Navarro, J.; Perezgrueso, A.; Barría, C.; Coll, M. Photo-identification as a tool to study small-spotted catshark Scyliorhinus canicula. J. Fish Biol. 2018, 92, 1657–1662. [Google Scholar] [CrossRef]
- Al-Jubouri, Q.; Al-Azawi, R.J.; Al-Taee, M.; Young, I. Efficient individual identification of zebrafish using Hue/Saturation/Value color model. Egypt. J. Aquat. Res. 2018, 44, 271–277. [Google Scholar] [CrossRef]
- Hirsch, P.E.; Eckmann, R. Individual identification of Eurasian perch Perca fluviatilis by means of their stripe patterns. Limnologica 2015, 54, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Huntingford, F.A.; Borçato, F.L.; Mesquita, F.O. Identifying individual common carp Cyprinus carpio using scale pattern. J. Fish Biol. 2013, 83, 1453–1458. [Google Scholar] [CrossRef] [PubMed]
- Stien, L.H.; Nilsson, J.; Bui, S.; Fosseidengen, J.E.; Kristiansen, T.S.; Øverli; Folkedal, O. Consistent melanophore spot patterns allow long-term individual recognition of Atlantic salmon Salmo salar. J. Fish Biol. 2017, 91, 1699–1712. [Google Scholar] [CrossRef] [Green Version]
- Pedersen, M.; Mohammed, A. Photo identification of individual salmo trutta based on deep learning. Appl. Sci. 2021, 11, 9039. [Google Scholar] [CrossRef]
- Hook, S.A.; McMurray, C.; Ripley, D.M.; Allen, N.; Moritz, T.; Grunow, B.; Shiels, H.A. Recognition software successfully aids the identification of individual small-spotted catsharks Scyliorhinus canicula during their first year of life. J. Fish Biol. 2019, 95, 1465–1470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castillo, G.C.; Sandford, M.E.; Hung, T.C.; Yang, W.R.; Tigan, G.; Ellison, L.; Lindberg, J.C.; Van Nieuwenhuyse, E.E. Evaluation of chromatophores as natural marks for delta smelt: The effects of life-stage and light intensity. Environ. Biol. Fishes 2019, 102, 1137–1147. [Google Scholar] [CrossRef] [Green Version]
- Bekkozhayeva, D.; Saberioon, M.; Cisar, P. Automatic individual non-invasive photo-identification of fish (Sumatra barb Puntigrus tetrazona) using visible patterns on a body. Aquac. Int. 2021, 29, 1481–1493. [Google Scholar] [CrossRef]
- Schraml, R.; Hofbauer, H.; Jalilian, E.; Bekkozhayeva, D.; Mohammadmehdi, S.; Cisar, P.; Uhl, A. Towards fish individuality-based aquaculture. IEEE Trans. Ind. Inform. 2020, 17, 4356–4366. [Google Scholar] [CrossRef]
- Cisar, P.; Bekkozhayeva, D.; Movchan, O.; Saberioon, M.; Schraml, R. Computer vision based individual fish identification using skin dot pattern. Sci. Rep. 2021, 11, 16904. [Google Scholar] [CrossRef] [PubMed]
- Ibáñez, A.L.; Guerra, E.; Pacheco-Almanzar, E. Fish Species Identification Using the Rhombic Squamation Pattern. Front. Mar. Sci. 2020, 7, 211. [Google Scholar] [CrossRef]
- Šonka, M.; Hlavac, V.; Boyle, R. Image Processing, Analysis, and Machine Vision; Springer: Berlin/Heidelberg, Germany, 2008; ISBN 978-0-495-08252-1. [Google Scholar]
- Dalal, N.; Triggs, B. Histograms of oriented gradients for human detection. In Proceedings of the 2005 IEEE computer society conference on computer vision and pattern recognition (CVPR’05), San Diego, CA, USA, 20–26 June 2005; Volume I, pp. 886–893. [Google Scholar] [CrossRef] [Green Version]
- Sfakianakis, D.G.; Katharios, P.; Tsirigotakis, N.; Doxa, C.K.; Kentouri, M. Lateral line deformities in wild and farmed sea bass (Dicentrarchuslabrax L.) and sea bream (Sparus aurata L.). J. Appl. Ichthyol. 2013, 29, 1015–1021. [Google Scholar] [CrossRef]
- Arechavala-Lopez, P.; Sanchez-Jerez, P.; Bayle-Sempere, J.T.; Sfakianakis, D.G.; Somarakis, S. Discriminating farmed gilthead sea bream Sparus aurataand European sea bass Dicentrarchus labrax from wild stocks through scales andotoliths. J. Fish Biol. 2012, 80, 2159–2175. [Google Scholar] [CrossRef]
- Ibáñez, A.L.; Gallardo-Cabello, M. Identification of two Mugilidae species, Mugil cephalus and M. curema (Pisces: Mugilidae), using the ctenii of their scales. Bull. Mar. Sci. 2005, 77, 305–307. [Google Scholar]
- Ibáñez, A.L. Fish scale shape variation by year and by geographic location, could scales be useful to trace fish? A case study on the Gulf of Mexico. Fish. Res. 2014, 156, 34–38. [Google Scholar] [CrossRef]
- Eldar, G.; Abbasali, A.; Sharafkxanim, A.; David, C. The Surface Fractal Structure of Fish Scales. Open J. Inorg. Non-metallic Mater. 2014, 4, 7–11. [Google Scholar] [CrossRef] [Green Version]
- Goodrich, E.S. The vertebrata craniata (cyclostomes and fishes). Ray E 1909, IX, 518. [Google Scholar]
- Ibañez, A.L.; Cowx, I.G.; O’Higgins, P. Geometric morphometric analysis of fish scales for identifying genera, species, and local populations within the Mugilidae. Can. J. Fish. Aquat. Sci. 2007, 64, 1091–1100. [Google Scholar] [CrossRef] [Green Version]
- Chervinski, J. Using scales for identification of four mugilidae species. Aquaculture 1984, 38, 79–81. [Google Scholar] [CrossRef]
- Chervinski, J. Identification of four tilapia species from Lake Kinneret, Israel, by the form of their scales. Aquaculture 1986, 52, 235–236. [Google Scholar] [CrossRef]
- Sato, T.; Tawa, A.; Sakuma, K.; Sakurai, M. Larval identification based on melanophore patterns in two Auxis species, bullet tuna Auxis rochei and frigate tuna Auxis thazard, from the northwest Pacific Ocean. Fish. Sci. 2020, 86, 625–631. [Google Scholar] [CrossRef]
- Bräger, Z.; Staszny, Á.; Mertzen, M.; Moritz, T.; Horváth, G. Fish scale identification: From individual to species-specific shape variability. Acta Ichthyol. Piscat. 2017, 47, 331–338. [Google Scholar] [CrossRef] [Green Version]
- Yusup, I.M.; Iqbal, M.; Jaya, I. Real-time reef fishes identification using deep learning. IOP Conf. Ser. Earth Environ. Sci. 2020, 429, 012046. [Google Scholar] [CrossRef]




| Region of Interest | Accuracy for ST (300 Fish) | Accuracy for LT (32 Fish) |
|---|---|---|
| ROI (LL) | 100% | 100% |
| ROI (O) | 91.66% | 40.62% |
| ROI (HLL) | 98.66% | 96.87% |
| ROI (S) | 98.66% | 93.75% |
| Data Collection | 01 | 02 | 03 | 04 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ROI1 | ROI2 | ROI3 | ROI1 | ROI2 | ROI3 | ROI1 | ROI2 | ROI3 | ROI1 | ROI2 | ROI3 | |
| 01 | 100 | 100 | 100 | 80.64 | 80.64 | 77.41 | 100 | 96.77 | 64.51 | 90.32 | 83.87 | 32.25 |
| 02 | 100 | 100 | 100 | 80.64 | 80.64 | 67.74 | 70.41 | 80.64 | 29.03 | |||
| 03 | 100 | 100 | 100 | 100 | 100 | 87.09 | ||||||
| 04 | 100 | 100 | 100 | |||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bekkozhayeva, D.; Cisar, P. Image-Based Automatic Individual Identification of Fish without Obvious Patterns on the Body (Scale Pattern). Appl. Sci. 2022, 12, 5401. https://doi.org/10.3390/app12115401
Bekkozhayeva D, Cisar P. Image-Based Automatic Individual Identification of Fish without Obvious Patterns on the Body (Scale Pattern). Applied Sciences. 2022; 12(11):5401. https://doi.org/10.3390/app12115401
Chicago/Turabian StyleBekkozhayeva, Dinara, and Petr Cisar. 2022. "Image-Based Automatic Individual Identification of Fish without Obvious Patterns on the Body (Scale Pattern)" Applied Sciences 12, no. 11: 5401. https://doi.org/10.3390/app12115401
APA StyleBekkozhayeva, D., & Cisar, P. (2022). Image-Based Automatic Individual Identification of Fish without Obvious Patterns on the Body (Scale Pattern). Applied Sciences, 12(11), 5401. https://doi.org/10.3390/app12115401






