BIOPEP-UWM Virtual—A Novel Database of Food-Derived Peptides with In Silico-Predicted Biological Activity
Abstract
:1. Introduction
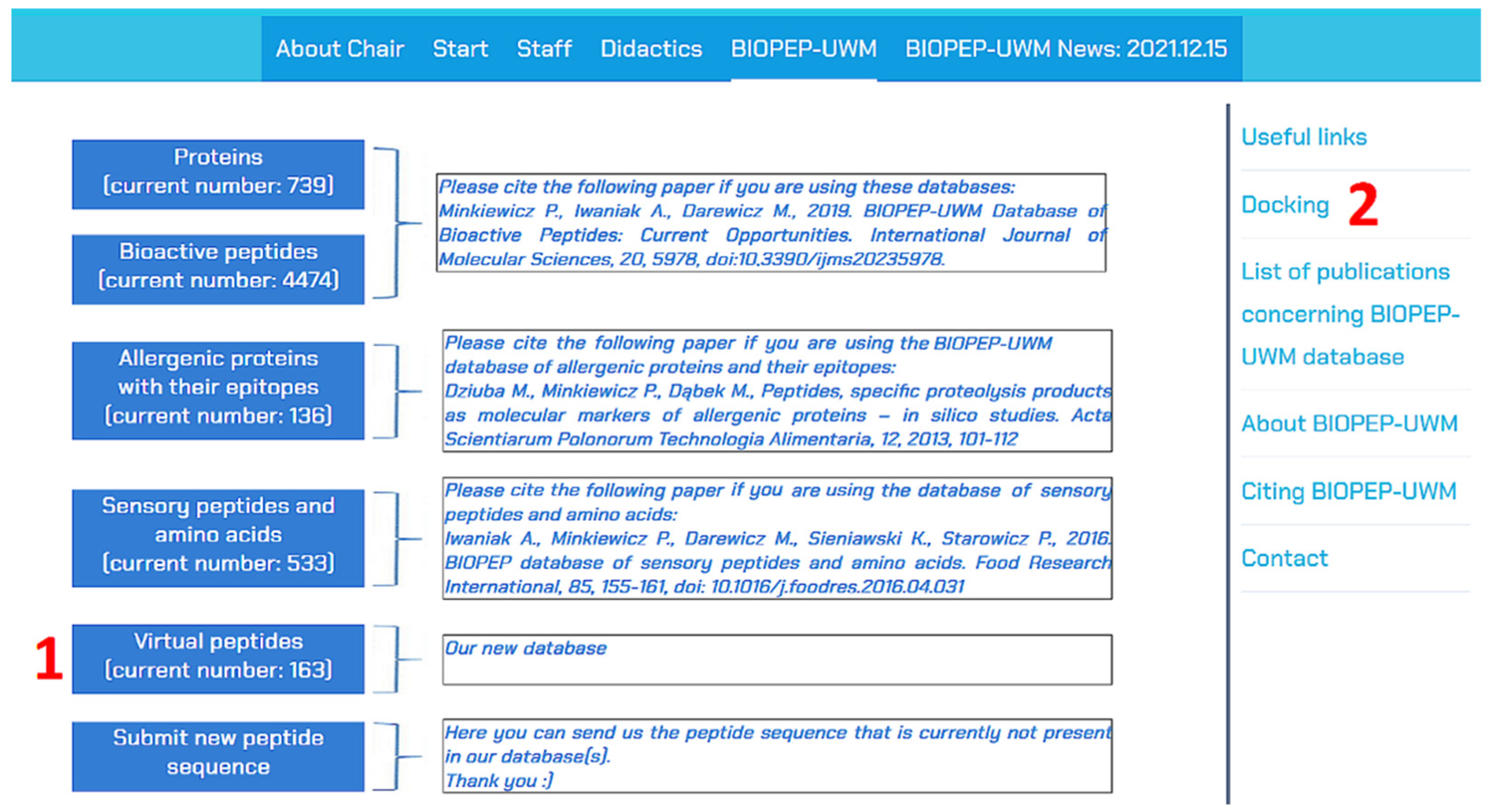
2. Database Description


- -
- name (if possible) and sequence;
- -
- function information—information about the predicted target biomacromolecule;
- -
- bibliographic data with the reference paper describing the peptide;
- -
- additional information, including the annotated peptide structure using chemical codes SMILES [17] and InChI [18], InChIKey identifier, specification of the method used for bioactivity prediction (to date, mainly molecular docking), information about other activities discovered experimentally or predicted using computational methods, and/or peptide taste (if available). The last category of content may include information about the main tastes (bitter, umami, sweet, sour, or salty) as well as enhancement or suppression of taste (e.g., bitterness-suppressing or umami-enhancing peptides). This information is taken from the BIOPEP-UWM database of sensory peptides and amino acids [15]. Information about the activity or taste of the peptide cites the database or databases providing this information;
- -
- a database reference tab providing information about compound annotations in other databases (if available). The set of databases cited in this tab is described in our previous article [12]. Information about peptide annotation includes database name and compound ID. This tab also contains information about the annotation of peptides in the BIOPEP-UWM database of bioactive peptides and/or the BIOPEP-UWM database of sensory peptides and amino acids;
- -
- entire peptide data arranged for printing, available via the “Screen and print peptide data”. A table screenshot of the content of the above tab is shown in Figure 4.
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Agyei, D.; Tsopmo, A.; Udenigwe, C.C. Bioinformatics and peptidomics approaches to the discovery and analysis of food-derived bioactive peptides. Anal. Bioanal. Chem. 2018, 410, 3463–3472. [Google Scholar] [CrossRef] [PubMed]
- Tu, M.; Cheng, S.; Lu, W.; Du, M. Advancement and prospects of bioinformatics analysis for studying bioactive peptides from food-derived protein: Sequence, structure, and functions. Trends Anal. Chem. 2018, 105, 7–17. [Google Scholar] [CrossRef]
- Iwaniak, A.; Darewicz, M.; Mogut, D.; Minkiewicz, P. Elucidation of the role of in silico methodologies in approaches to studying bioactive peptides derived from foods. J. Funct. Foods 2019, 61, 103486. [Google Scholar] [CrossRef]
- Yao, Y.; Luo, Z.; Zhang, X. In silico evaluation of marine fish proteins as nutritional supplement for COVID-2019 patients. Food Funct. 2020, 11, 5565–5572. [Google Scholar] [CrossRef]
- Heydari, H.; Golmohammadi, R.; Mirnejad, R.; Tebyanian, H.; Fasihi-Ramandi, M.; Moghaddam, M.M. Antiviral peptides against Coronaviridae family: A review. Peptides 2021, 139, 170526. [Google Scholar] [CrossRef]
- Zhao, W.; Li, W.; Yu, Z.; Wu, S.; Ding, L.; Liu, J. Identification of lactoferrin-derived peptides as potential inhibitors against the main protease of SARS-CoV-2. LWT 2022, 154, 112684. [Google Scholar] [CrossRef]
- Yu, Z.; Wang, Y.; Zhao, W.; Li, J.; Shuian, D.; Liu, J. Identification of Oncorhynchus mykiss nebulin-derived peptides as bitter taste receptor TAS2R14 blockers by in silico screening and molecular docking. Food Chem. 2022, 368, 130839. [Google Scholar] [CrossRef]
- Li, C.; Mora, L.; Toldrá, F. Structure-function relationship of small peptides generated during the ripening of Spanish dry-cured ham: Peptidome, molecular stability and computational modelling. Food Chem. 2022, 375, 131673. [Google Scholar] [CrossRef]
- Vidal-Limon, A.; Aguilar-Toalá, J.E.; Liceaga, A.M. Integration of molecular docking analysis and molecular dynamics simulations for studying food proteins and bioactive peptides. J. Agric. Food Chem. 2022, 70, 934–943. [Google Scholar] [CrossRef]
- Khan, A.T.; Chowdhury, G.M.; Hafsah, J.; Maruf, M.; Raihan, M.R.; Chowdhury, M.T.; Nawal, N.; Tasnim, N.; Saha, P.; Roy, P.; et al. A student led computational screening of peptide inhibitors against main protease of SARS-CoV-2. Biochem. Mol. Biol. Educ. 2022, 50, 7–20. [Google Scholar] [CrossRef]
- Medina-Franco, J.L.; Martinez-Mayorga, K.; Fernández-de Gortari, E.; Kirchmair, J.; Bajorath, J. Rationality over fashion and hype in drug design. F1000Research 2021, 10, 397. [Google Scholar] [CrossRef] [PubMed]
- Minkiewicz, P.; Iwaniak, A.; Darewicz, M. BIOPEP-UWM database of bioactive peptides: Current opportunities. Int. J. Mol. Sci. 2019, 20, 5978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwaniak, A.; Darewicz, M.; Minkiewicz, P. Chapter 14—Databases of Bioactive Peptides. In Biologically Active Peptides. From Basic Science to Applications for Human Health; Toldrá, F., Wu, J., Eds.; Elsevier Inc.: Oxford, UK, 2021; pp. 309–330. [Google Scholar] [CrossRef]
- Dziuba, M.; Minkiewicz, P.; Dąbek, M. Peptides, specific proteolysis products as molecular markers of allergenic proteins—In Silico studies. Acta Sci. Pol. Technol. Aliment. 2013, 12, 101–112. Available online: https://www.food.actapol.net/volume12/issue1/abstract-10.html (accessed on 13 July 2022). [PubMed]
- Iwaniak, A.; Minkiewicz, P.; Darewicz, M.; Sieniawski, K.; Starowicz, P. BIOPEP database of sensory peptides and amino acids. Food Res. Int. 2016, 85, 155–161. [Google Scholar] [CrossRef]
- Wenhui, T.; Shumin, H.; Yongliang, Z.; Liping, S.; Hua, Y. Identification of in vitro angiotensin-converting enzyme and dipeptidyl peptidase IV inhibitory peptides from draft beer by virtual screening and molecular docking. J. Sci. Food Agric. 2022, 102, 1085–1094. [Google Scholar] [CrossRef]
- Weininger, D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 1988, 28, 31–36. [Google Scholar] [CrossRef]
- Goodman, J.M.; Pletnev, I.; Thiessen, P.; Bolton, E.; Heller, S.R. InChI version 1.06: Now more than 99.99% reliable. J. Cheminform. 2021, 13, 40. [Google Scholar] [CrossRef]
- PDBe-KB Consortium. PDBe-KB: Collaboratively defining the biological context of structural data. Nucleic Acids Res. 2022, 50, D534–D542. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Barrett, A.J.; Thomas, P.D.; Huang, X.; Bateman, A.; Finn, R.D. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018, 46, D624–D632. [Google Scholar] [CrossRef]
- Hähnke, V.D.; Kim, S.; Bolton, E.E. PubChem chemical structure standardization. J. Cheminform. 2018, 10, 36. [Google Scholar] [CrossRef]
- Minkiewicz, P.; Darewicz, M.; Iwaniak, A.; Turło, M. Proposal of the annotation of phosphorylated amino acids and peptides using biological and chemical codes. Molecules 2021, 26, 712. [Google Scholar] [CrossRef]
- Minkiewicz, P.; Iwaniak, A.; Darewicz, M. Using internet databases for food science organic chemistry students to discover chemical compound information. J. Chem. Educ. 2015, 92, 874–876. [Google Scholar] [CrossRef]
- Panyayai, T.; Ngamphiw, C.; Tongsima, S.; Mhuantong, W.; Limsripraphan, W.; Choowongkomon, K.; Sawatdichaikul, O. FeptideDB: A web application for new bioactive peptides from food protein. Heliyon 2019, 5, e02076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Grimm, M.; Dai, W.; Hou, M.; Xiao, Z.-X.; Cao, Y. CB-Dock: A web server for cavity detection-guided protein–ligand blind docking. Acta Pharmacol. Sin. 2020, 41, 138–144. [Google Scholar] [CrossRef]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro web server for protein-protein docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Minkiewicz, P.; Iwaniak, A.; Darewicz, M. BIOPEP-UWM Virtual—A Novel Database of Food-Derived Peptides with In Silico-Predicted Biological Activity. Appl. Sci. 2022, 12, 7204. https://doi.org/10.3390/app12147204
Minkiewicz P, Iwaniak A, Darewicz M. BIOPEP-UWM Virtual—A Novel Database of Food-Derived Peptides with In Silico-Predicted Biological Activity. Applied Sciences. 2022; 12(14):7204. https://doi.org/10.3390/app12147204
Chicago/Turabian StyleMinkiewicz, Piotr, Anna Iwaniak, and Małgorzata Darewicz. 2022. "BIOPEP-UWM Virtual—A Novel Database of Food-Derived Peptides with In Silico-Predicted Biological Activity" Applied Sciences 12, no. 14: 7204. https://doi.org/10.3390/app12147204
APA StyleMinkiewicz, P., Iwaniak, A., & Darewicz, M. (2022). BIOPEP-UWM Virtual—A Novel Database of Food-Derived Peptides with In Silico-Predicted Biological Activity. Applied Sciences, 12(14), 7204. https://doi.org/10.3390/app12147204








